Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein - PubMed (original) (raw)
Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein
X Ma et al. Proc Natl Acad Sci U S A. 1996.
Abstract
In the current model for bacterial cell division, FtsZ protein forms a ring that marks the division plane, creating a cytoskeletal framework for the subsequent action of other proteins such as FtsA. This putative protein complex ultimately generates the division septum. Herein we report that FtsZ and FtsA proteins tagged with green fluorescent protein (GEP) colocalize to division-site ring-like structures in living bacterial cells in a visible space between the segregated nucleoids. Cells with higher levels of FtsZ-GFP or with FtsA-GFP plus excess wild-type FtsZ were inhibited for cell division and often exhibited bright fluorescent spiral tubules that spanned the length of the filamentous cells. This suggests that FtsZ may switch from a septation-competent localized ring to an unlocalized spiral under some conditions and that FtsA can bind to FtsZ in both conformations. FtsZ-GFP also formed nonproductive but localized aggregates at a higher concentration that could represent FtsZ nucleation sites. The general domain structure of FtsZ-GFP resembles that of tubulin, since the C terminus of FtsZ is not required for polymerization but may regulate polymerization state. The N-terminal portion of Rhizobium FtsZ polymerized in Escherichia coli and appeared to copolymerize with E. coli FtsZ, suggesting a degree of interspecies functional conservation. Analysis of several deletions of FtsA-GFP suggests that multiple segments of FtsA are important for its localization to the FtsZ ring.
Figures
Figure 1
Plasmids expressing fusion proteins. Diagrammed below the wild-type E. coli FtsZ and FtsA and R. meliloti FtsZ1 protein diagrams are the various GFP fusions and deletions, all present on either pBluescript SK+ or pBC SK+. Numbers refer to the amino acid residue of FtsZ or FtsA at relevant junctions. Open boxes for FtsZ represent the conserved N terminus and for FtsA represent the entire encoded protein. Solid boxes denote the variable C terminus of FtsZ (residues 317–383 in_E. coli_ FtsZ). Dark shaded boxes denote GFP and light shaded boxes denote the N-terminal 20 amino acids of vector_lacZ_ from the start codon to the_Sac_I site. Hatched boxes represent the divergent and unusually long C terminus of R. meliloti FtsZ1.
Figure 2
FtsZ–GFP forms rings at midpoints of E. coli cells. Cells in A–D were viewed by conventional fluorescence; cells in E and_F_ were imaged by deconvolution. All were from colonies on plates without IPTG. (A) JM109/pZG, showing localization to cell midpoints. Arrows point to double bands. (B and C) FtsZ–GFP fluorescence (JM109/pZG) in cells undergoing visible septation. (Upper) With fluorescence/phase-contrast. (Lower) Phase-contrast only. (D) JM109 containing pCSK100 (GFP only). (E) JM109/pZG cell with FtsZ–GFP ring. Upper and Lower are two different viewing angles of the same cell. (F) Same as_E_ except with incomplete fluorescent ring. (Bars:A and D, 5 μm; B, C, E, and_F_, 1 μm; bar shown in E only.)
Figure 3
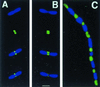
FtsZ–GFP localizes to internucleoid regions. (A and B) Individual cells of JM109/pZG stained with DAPI (1 μg/ml), viewed for DAPI fluorescence only (top part), GFP fluorescence only (second part from top), and DAPI + GFP (lower two parts, composite images, with lowest part darkened to improve visualization of the internucleoid space). (C) Filamentous cell (JM109/pZG) in a population of mostly normal cells, imaged for both DAPI and GFP. (Bar = 1 μm.)
Figure 4
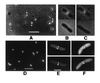
Tubules, bands, and dots in cells with FtsZ–GFP. JM109/pZG cells were from colonies on plates without IPTG. (A) Moderately filamentous cell showing regularly spaced bands. (B) Filamentous cell in a population of mostly normal cells, showing regularly spaced dots. (C) Nonfilamentous cell containing spiral polymers, imaged with deconvolution. (D) Same cell as in C but from a different angle. (E) Filamentous cell with spiral polymers. (F) Filamentous cells showing both spiral polymers and regularly spaced dots. (Bars: A, B, E, and_F_, 5 μm; C and D, 1 μm.)
Figure 5
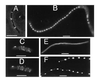
FtsA–GFP localization. (A–E) Cells with pAG and extra copies of _lacI_q, from colonies on plates without IPTG. (A) JM109/pAG cells with FtsA–GFP fluorescence localized to cell midpoints. Arrows point to double bands. (B) MC1061/pAG cells, with_lacI_q cloned into the plasmid. (C) Deconvolution image of JM109/pAG cells, one showing FtsA–GFP localization to a midcell ring, the other showing spiral structures. (D) Same cells as in C, at a different angle. (E) Deconvolution image of JM109/pAG cell showing midcell fluorescence and spiral structures. (F) Filamentous JM109/pAG cells with no extra copies of_lacI_q; higher levels of FtsA–GFP cause bulges and more diffuse fluorescence. (G) Filamentous JM105/pAG/pMK4 cell expressing FtsA–GFP and wild-type FtsZ, with IPTG. (Bars: A, B, F, and G, 5 μm;C and D, 1 μm.)
Figure 6
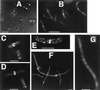
Effects of deletions of GFP-tagged FtsA and FtsZ. (A) JM109/pZGCΔ cell showing typical large unlocalized structures. (B) JM109/pRmZG cells with axial polymers produced by R. meliloti FtsZ1Δ-GFP. Arrows point to midcell localization observed in a subset of cells. (C) JM105/pZGNΔ cells showing uniform fluorescence; the regions of bright fluorescence at the poles seen in some cells are large inclusion bodies. (D) JM105/pAGCΔ. (E) JM105/pAGNΔ1. (F) JM105/pAGNΔ2 cells; cell surface fluorescence is especially apparent. (Bars = 1 μm.)
Similar articles
- Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring.
Ma X, Sun Q, Wang R, Singh G, Jonietz EL, Margolin W. Ma X, et al. J Bacteriol. 1997 Nov;179(21):6788-97. doi: 10.1128/jb.179.21.6788-6797.1997. J Bacteriol. 1997. PMID: 9352931 Free PMC article. - Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain.
Yu XC, Tran AH, Sun Q, Margolin W. Yu XC, et al. J Bacteriol. 1998 Mar;180(5):1296-304. doi: 10.1128/JB.180.5.1296-1304.1998. J Bacteriol. 1998. PMID: 9495771 Free PMC article. - FtsZ dynamics during the division cycle of live Escherichia coli cells.
Sun Q, Margolin W. Sun Q, et al. J Bacteriol. 1998 Apr;180(8):2050-6. doi: 10.1128/JB.180.8.2050-2056.1998. J Bacteriol. 1998. PMID: 9555885 Free PMC article. - Building the Bacterial Divisome at the Septum.
Morrison JJ, Camberg JL. Morrison JJ, et al. Subcell Biochem. 2024;104:49-71. doi: 10.1007/978-3-031-58843-3_4. Subcell Biochem. 2024. PMID: 38963483 Review. - The keepers of the ring: regulators of FtsZ assembly.
Ortiz C, Natale P, Cueto L, Vicente M. Ortiz C, et al. FEMS Microbiol Rev. 2016 Jan;40(1):57-67. doi: 10.1093/femsre/fuv040. Epub 2015 Sep 15. FEMS Microbiol Rev. 2016. PMID: 26377318 Review.
Cited by
- The price of tags in protein localization studies.
Margolin W. Margolin W. J Bacteriol. 2012 Dec;194(23):6369-71. doi: 10.1128/JB.01640-12. Epub 2012 Sep 7. J Bacteriol. 2012. PMID: 22961859 Free PMC article. No abstract available. - Interaction between cell division proteins FtsE and FtsZ.
Corbin BD, Wang Y, Beuria TK, Margolin W. Corbin BD, et al. J Bacteriol. 2007 Apr;189(8):3026-35. doi: 10.1128/JB.01581-06. Epub 2007 Feb 16. J Bacteriol. 2007. PMID: 17307852 Free PMC article. - Timing of FtsZ assembly in Escherichia coli.
Den Blaauwen T, Buddelmeijer N, Aarsman ME, Hameete CM, Nanninga N. Den Blaauwen T, et al. J Bacteriol. 1999 Sep;181(17):5167-75. doi: 10.1128/JB.181.17.5167-5175.1999. J Bacteriol. 1999. PMID: 10464184 Free PMC article. - Exploring intracellular space: function of the Min system in round-shaped Escherichia coli.
Corbin BD, Yu XC, Margolin W. Corbin BD, et al. EMBO J. 2002 Apr 15;21(8):1998-2008. doi: 10.1093/emboj/21.8.1998. EMBO J. 2002. PMID: 11953319 Free PMC article. - Green fluorescent protein functions as a reporter for protein localization in Escherichia coli.
Feilmeier BJ, Iseminger G, Schroeder D, Webber H, Phillips GJ. Feilmeier BJ, et al. J Bacteriol. 2000 Jul;182(14):4068-76. doi: 10.1128/JB.182.14.4068-4076.2000. J Bacteriol. 2000. PMID: 10869087 Free PMC article.
References
- Rothfield L I, Zhao C-R. Cell. 1996;84:183–186. - PubMed
- Chang F, Nurse P. Cell. 1996;84:191–194. - PubMed
- Donachie W D. Annu Rev Microbiol. 1993;47:199–230. - PubMed
- Lutkenhaus J. Mol Microbiol. 1993;9:403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases