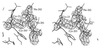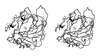Crystal structure of the secretory form of membrane-associated human carbonic anhydrase IV at 2.8-A resolution - PubMed (original) (raw)
Crystal structure of the secretory form of membrane-associated human carbonic anhydrase IV at 2.8-A resolution
T Stams et al. Proc Natl Acad Sci U S A. 1996.
Abstract
It has recently been demonstrated that the C-terminal deletion mutant of recombinant human carbonic anhydrase IV (G267X CA IV) converts the normally glycosylphosphatidylinositol-anchored enzyme into a soluble secretory form which has the same catalytic properties as the membrane-associated enzyme purified from human tissues. We have determined the three-dimensional structure of the secretory form of human CA IV by x-ray crystallographic methods to a resolution of 2.8 A. Although the zinc binding site and the hydrophobic substrate binding pocket of CA IV are generally similar to those of other mammalian isozymes, unique structural differences are found elsewhere in the active site. Two disufide linkages, Cys-6-Cys-11G and Cys-23-Cys-203, stabilize the conformation of the N-terminal domain. The latter disulfide additionally stabilizes an active site loop containing a cis-peptide linkage between Pro-201 and Thr-202 (this loop contains catalytic residue Thr-199). On the opposite side of the active site, the Val-131-Asp-136 segment adopts an extended loop conformation instead of an alpha-helix conformation as found in other isozymes. Finally, the C terminus is surrounded by a substantial electropositive surface potential, which is likely to stabilize the interaction of CA IV with the negatively charged phospholipid headgroups of the membrane. These structural features are unique to CA IV and provide a framework for the design of sulfonamide inhibitors selective for this particular isozyme.
Figures
Figure 1
Sequence alignment of CA IV and CA II based on superposition of their three-dimensional structures. Relative to CA II, CA IV contains 8 insertions and 3 deletions; the overall sequence identity is 33%. The CA IV numbering scheme used in this work is based on this alignment; inserted residues are indicated by the number of the residue preceding the insertion and the suffix A, B, C, etc.
Figure 2
Omit map of the zinc binding site of CA IV contoured at 4σ; refined atomic coordinates are superimposed and selected active site residues are labeled. Zinc is tetrahedrally coordinated by His-94, His-96, His-119, and a sulfate oxygen. The hydrophobic substrate binding pocket is visible in the upper right background.
Figure 3
Omit map showing the Cys-23–Cys-203 disulfide linkage and the Pro-201–Thr-202 _cis_-peptide linkage (contoured at 4σ). Refined atomic coordinates are superimposed and selected active site residues are labeled.
Figure 4
Least-squares Cα superposition of CA IV (thick bonds) and CA II (thin bonds). The active site zinc ion appears as a gray sphere, and disulfide linkages of CA IV are indicated by large black spheres. Note that although the overall folds of the two isozymes are generally similar, there are significant differences in the region of residue 131 (small black sphere). The extended conformation of the Arg-129–Asn-130 linkage in CA IV may enhance its susceptibility to proteolysis (17, 23).
Figure 5
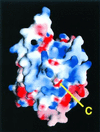
Electrostatic surface potential of CA IV calculated with
grasp
(55); the color scale ranges from −8_kT_ (red) to +8 kT (blue). Note the extensive positive electrostatic surface potential surrounding the C terminus. This feature reflects the adaptation of the carbonic anhydrase framework for GPI-anchoring and association with the membrane.
Figure 6
Cartoon of the CA IV–membrane interaction. The CA IV isozyme is anchored to the membrane by a GPI tail attached to its C terminus (yellow), which orients the enzyme active site toward the lumen for catalysis. This orientation is further stabilized by the interactions of 11 arginine, lysine, and histidine residues flanking the C terminus with the negatively charged phospholipid headgroups (red) of the membrane. The active site zinc ion appears as a white sphere, and the two disulfide linkages are indicated by bonded yellow spheres. The figure was prepared with
molscript
(56) and
raster3D
(57, 58).
Similar articles
- Expression, assay, and structure of the extracellular domain of murine carbonic anhydrase XIV: implications for selective inhibition of membrane-associated isozymes.
Whittington DA, Grubb JH, Waheed A, Shah GN, Sly WS, Christianson DW. Whittington DA, et al. J Biol Chem. 2004 Feb 20;279(8):7223-8. doi: 10.1074/jbc.M310809200. Epub 2003 Dec 3. J Biol Chem. 2004. PMID: 14660577 - Crystal structure of the dimeric extracellular domain of human carbonic anhydrase XII, a bitopic membrane protein overexpressed in certain cancer tumor cells.
Whittington DA, Waheed A, Ulmasov B, Shah GN, Grubb JH, Sly WS, Christianson DW. Whittington DA, et al. Proc Natl Acad Sci U S A. 2001 Aug 14;98(17):9545-50. doi: 10.1073/pnas.161301298. Epub 2001 Aug 7. Proc Natl Acad Sci U S A. 2001. PMID: 11493685 Free PMC article. - Structures of murine carbonic anhydrase IV and human carbonic anhydrase II complexed with brinzolamide: molecular basis of isozyme-drug discrimination.
Stams T, Chen Y, Boriack-Sjodin PA, Hurt JD, Liao J, May JA, Dean T, Laipis P, Silverman DN, Christianson DW. Stams T, et al. Protein Sci. 1998 Mar;7(3):556-63. doi: 10.1002/pro.5560070303. Protein Sci. 1998. PMID: 9541386 Free PMC article. - X-ray crystallographic studies of mammalian carbonic anhydrase isozymes.
Stams T, Christianson DW. Stams T, et al. EXS. 2000;(90):159-74. doi: 10.1007/978-3-0348-8446-4_9. EXS. 2000. PMID: 11268515 Review. No abstract available. - Recent advances in structural studies of the carbonic anhydrase family: the crystal structure of human CA IX and CA XIII.
Supuran CT, Di Fiore A, Alterio V, Monti SM, De Simone G. Supuran CT, et al. Curr Pharm Des. 2010;16(29):3246-54. doi: 10.2174/138161210793429841. Curr Pharm Des. 2010. PMID: 20819069 Review.
Cited by
- Roles of the conserved aspartate and arginine in the catalytic mechanism of an archaeal beta-class carbonic anhydrase.
Smith KS, Ingram-Smith C, Ferry JG. Smith KS, et al. J Bacteriol. 2002 Aug;184(15):4240-5. doi: 10.1128/JB.184.15.4240-4245.2002. J Bacteriol. 2002. PMID: 12107142 Free PMC article. - Insights into the role of reactive sulfhydryl groups of Carbonic Anhydrase III and VII during oxidative damage.
Monti DM, De Simone G, Langella E, Supuran CT, Di Fiore A, Monti SM. Monti DM, et al. J Enzyme Inhib Med Chem. 2017 Dec;32(1):5-12. doi: 10.1080/14756366.2016.1225046. Epub 2016 Oct 21. J Enzyme Inhib Med Chem. 2017. PMID: 27766895 Free PMC article. Review. - Structural analysis of charge discrimination in the binding of inhibitors to human carbonic anhydrases I and II.
Srivastava DK, Jude KM, Banerjee AL, Haldar M, Manokaran S, Kooren J, Mallik S, Christianson DW. Srivastava DK, et al. J Am Chem Soc. 2007 May 2;129(17):5528-37. doi: 10.1021/ja068359w. Epub 2007 Apr 4. J Am Chem Soc. 2007. PMID: 17407288 Free PMC article. - Alpha-Carbonic Anhydrases from Hydrothermal Vent Sources as Potential Carbon Dioxide Sequestration Agents: In Silico Sequence, Structure and Dynamics Analyses.
Manyumwa CV, Emameh RZ, Tastan Bishop Ö. Manyumwa CV, et al. Int J Mol Sci. 2020 Oct 29;21(21):8066. doi: 10.3390/ijms21218066. Int J Mol Sci. 2020. PMID: 33138066 Free PMC article. - Stabilization of anionic and neutral forms of a fluorophoric ligand at the active site of human carbonic anhydrase I.
Manokaran S, Banerjee J, Mallik S, Srivastava DK. Manokaran S, et al. Biochim Biophys Acta. 2010 Oct;1804(10):1965-73. doi: 10.1016/j.bbapap.2010.06.024. Epub 2010 Jul 8. Biochim Biophys Acta. 2010. PMID: 20620244 Free PMC article.
References
- Tashian R E. Adv Genet. 1992;30:321–356. - PubMed
- Sly W S, Hu P Y. Annu Rev Biochem. 1995;64:375–401. - PubMed
- Hewett-Emmett D, Tashian R E. Mol Phylogenet Evol. 1996;5:50–77. - PubMed
- Coleman J E. In: Zinc Enzymes. Bertini I, Luchinat C, Maret W, Zeppezauer M, editors. Boston: Birkhauser; 1986. pp. 49–58.
- Silverman D N, Lindskog S. Acc Chem Res. 1988;21:30–36.
Publication types
MeSH terms
Substances
Grants and funding
- GM34182/GM/NIGMS NIH HHS/United States
- R01 DK040163/DK/NIDDK NIH HHS/United States
- DK40163/DK/NIDDK NIH HHS/United States
- R01 GM034182/GM/NIGMS NIH HHS/United States
- GM45614/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases