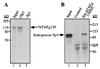Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100 - PubMed (original) (raw)
Comparative Study
Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100
N Tanese et al. Proc Natl Acad Sci U S A. 1996.
Abstract
Transcription factor TFIID is a multiprotein complex composed of the TATA box-binding protein (TBP) and multiple TBP-associated factors (TAFs). TFIID plays an essential role in mediating transcriptional activation by gene-specific activators. Numerous transcriptional activators have been characterized from mammalian cells; however, molecular analysis of the components of mammalian TFIID has been incomplete. Here we describe isolation of cDNAs encoding two TAF subunits of the human transcription factor TFIID. The first cDNA is predicted to encode the C-terminal 947 residues of the 130-kDa human TAF subunit, hTAFII130. The second cDNA encodes the C-terminal 801 residues of the 100-kDa subunit, hTAFII100. Recombinant TAFs expressed in human cells by transient transfections are capable of associating with the endogenous TAFs and TBP to form a TFIID complex in vivo. Protein binding experiments demonstrate that hTAFII130, like its Drosophila homolog dTAFII110, interacts with the glutamine-rich activation domains of the human transcription factor Sp1. Furthermore, hTAFII130 shows reduced binding to the Sp1 mutants with impaired ability to activate transcription, suggesting a role for hTAFII130 as a direct coactivator target for Sp1.
Figures
Figure 2
(A) Predicted C-terminal 947 amino acid sequence of hTAFII130. Peptides obtained from microsequencing of immunopurified hTAFII130 protein are underlined. (B) Schematic comparison of hTAFII130 with Drosophila dTAFII110. Region I (residues 449–528): 68% sequence similarity (46% identity) over 80 residues. Region II (residues 689–947): 72% similarity (55% identity) over 259 residues as determined by the
bestfit
sequence analysis program (Genetics Computer Group, Madison, WI). Glutamine-rich regions (presence of 20–30% glutamines in hTAFII130) are shaded and denoted by Q. hTAFII130N/C subdomain used in protein binding assays is indicated by a bracket. Dotted box represents the predicted missing N-terminal region. (C) Amino acid sequence alignment of conserved residues within regions I and II of dTAFII110 and hTAFII130.
Figure 3
Predicted C-terminal 801 amino acid sequence of hTAFII100. Peptide sequences obtained from microsequencing the endogenous hTAFII100 are indicated above the amino acid sequence. The dotted line below residues 557–611 represents 55 amino acids missing from a spliced variant of hTAFII100 isolated from the NTera cDNA library. The six WD repeats are indicated by the dotted arrows.
Figure 1
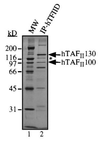
TFIID complex immunopurified from HeLa cells. hTAFIIs and hTBP were separated by SDS/PAGE and stained with silver. Positions of hTAFII130 and hTAFII100 are indicated. A 110-kDa hTAFII protein immunologically related to hTAFII130 is indicated by an asterisk (∗). We have obtained one peptide from this protein whose sequence is found in the predicted amino acid sequence of hTAFII130, suggesting that the 110-kDa hTAFII is a derivative or a breakdown product of hTAFII130.
Figure 4
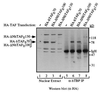
Recombinant HA-tagged hTAFIIs can be stably incorporated into the TFIID complex in vivo. Nuclear extracts were prepared from 293 cells transfected with a control plasmid (lanes 1 and 5) or expression plasmids encoding HA-hTAFII70 (lanes 2 and 6), HA-ΔNhTAFII100 containing C-terminal 704 residues (lanes 3 and 7), HA-ΔNhTAFII130 (C-terminal 947 residues, lanes 4 and 8) for 48 hours and TFIID was immunopurified using a polyclonal α-hTBP antibody. Presence of each recombinant hTAFII was detected by Western blotting using the α-HA antibody. Presence of endogenous hTAFIIs in immunopurified TFIID was detected by probing the same blot with a mixture of α-hTAFII monoclonal antibodies (data not shown).
Figure 5
Interactions between Sp1 and hTAFII130 detected in vitro. (A) Radiolabeled hTAFII130 was incubated with purified Sp1 bound to a DNA-affinity resin (lane 2) or a control resin prepared without Sp1 protein (lane 3). Input lane represents approximately 10% of each reaction. (B) Endogenous Sp1 was detected by Western blotting after incubating a crude HeLa cell nuclear extract (1.5 mg) with a resin containing immobilized HA-hTAFII130N/C, expressed and purified from E. coli (lane 3), and a control resin (lane 2).
Figure 6
(A) Sp1-hTAFII130 interaction detected by yeast two-hybrid assay. hTAFII130N/C fused to the GAL4 DNA binding domain (residues 1–147, lightly shaded) is shown schematically. β-Galactosidase activity measured from lysates of yeast cotransformed with AAD fusions are indicated on the right. (B) hTAFII130 interacts preferentially with the C-terminal subdomain of Sp1 B that activates transcription in HeLa cells. Yeast plasmids containing the full-length Sp1 B domain (amino acids 263–542), or the subdivisions Bn (263-424), or Bc (421-542) fused to the acidic activation domain (8) were cotransformed with lexADBD-hTAFII130N/C fusion. The resulting β-galactosidase activity measured relative to the activity of the full-length Sp1 B, is indicated on the right. ∗, Previously reported relative values of transcriptional activation by equivalent fusion constructs expressed and assayed in HeLa cells (8). (C) A linker substitution mutation (indicated by the black bar) in the C-terminal subdomain of Sp1 B reduces both interaction with hTAFII130 and transcriptional activation in human cells. The yeast two-hybrid experiments were conducted as described for_B_. ∗∗, Transcriptional activation values measured in 293 cells by transient transfection were taken from (31) and S. Smale (personal communication).
Similar articles
- Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators.
Chiang CM, Roeder RG. Chiang CM, et al. Science. 1995 Jan 27;267(5197):531-6. doi: 10.1126/science.7824954. Science. 1995. PMID: 7824954 - Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIF beta (RAP30) and incorporation into the TFIID complex.
Dubrovskaya V, Lavigne AC, Davidson I, Acker J, Staub A, Tora L. Dubrovskaya V, et al. EMBO J. 1996 Jul 15;15(14):3702-12. EMBO J. 1996. PMID: 8758937 Free PMC article. - Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators.
Saluja D, Vassallo MF, Tanese N. Saluja D, et al. Mol Cell Biol. 1998 Oct;18(10):5734-43. doi: 10.1128/MCB.18.10.5734. Mol Cell Biol. 1998. PMID: 9742090 Free PMC article. - Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators.
Chen JL, Attardi LD, Verrijzer CP, Yokomori K, Tjian R. Chen JL, et al. Cell. 1994 Oct 7;79(1):93-105. doi: 10.1016/0092-8674(94)90403-0. Cell. 1994. PMID: 7923382 - Mechanisms of transcriptional activation and repression can both involve TFIID.
Manley JL, Um M, Li C, Ashali H. Manley JL, et al. Philos Trans R Soc Lond B Biol Sci. 1996 Apr 29;351(1339):517-26. doi: 10.1098/rstb.1996.0050. Philos Trans R Soc Lond B Biol Sci. 1996. PMID: 8735274 Review.
Cited by
- Role of TAF4 in transcriptional activation by Rta of Epstein-Barr Virus.
Yang YC, Chang LK. Yang YC, et al. PLoS One. 2013;8(1):e54075. doi: 10.1371/journal.pone.0054075. Epub 2013 Jan 10. PLoS One. 2013. PMID: 23326574 Free PMC article. - Interaction between intrinsically disordered regions in transcription factors Sp1 and TAF4.
Hibino E, Inoue R, Sugiyama M, Kuwahara J, Matsuzaki K, Hoshino M. Hibino E, et al. Protein Sci. 2016 Nov;25(11):2006-2017. doi: 10.1002/pro.3013. Epub 2016 Aug 24. Protein Sci. 2016. PMID: 27515574 Free PMC article. - Prodos is a conserved transcriptional regulator that interacts with dTAF(II)16 in Drosophila melanogaster.
Hernández-Hernández A, Ferrús A. Hernández-Hernández A, et al. Mol Cell Biol. 2001 Jan;21(2):614-23. doi: 10.1128/MCB.21.2.614-623.2001. Mol Cell Biol. 2001. PMID: 11134347 Free PMC article. - Ovarian granulosa cell survival and proliferation requires the gonad-selective TFIID subunit TAF4b.
Voronina E, Lovasco LA, Gyuris A, Baumgartner RA, Parlow AF, Freiman RN. Voronina E, et al. Dev Biol. 2007 Mar 15;303(2):715-26. doi: 10.1016/j.ydbio.2006.12.011. Epub 2006 Dec 9. Dev Biol. 2007. PMID: 17207475 Free PMC article. - Wnt signaling targets ETO coactivation domain of TAF4/TFIID in vivo.
Wright KJ, Tjian R. Wright KJ, et al. Proc Natl Acad Sci U S A. 2009 Jan 6;106(1):55-60. doi: 10.1073/pnas.0811914106. Epub 2008 Dec 30. Proc Natl Acad Sci U S A. 2009. PMID: 19116271 Free PMC article.
References
- Roeder R G. Trends Biochem Sci. 1991;16:402–408. - PubMed
- Tjian R, Maniatis T. Cell. 1994;77:5–8. - PubMed
- Zawel L, Reinberg D. Annu Rev Biochem. 1995;64:533–561. - PubMed
- Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Cell. 1994;79:93–105. - PubMed
- Hoey T, Weinzierl R O, Gill G, Chen J L, Dynlacht B D, Tjian R. Cell. 1993;72:247–260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials