Characterization of individual polynucleotide molecules using a membrane channel - PubMed (original) (raw)
Characterization of individual polynucleotide molecules using a membrane channel
J J Kasianowicz et al. Proc Natl Acad Sci U S A. 1996.
Abstract
We show that an electric field can drive single-stranded RNA and DNA molecules through a 2.6-nm diameter ion channel in a lipid bilayer membrane. Because the channel diameter can accommodate only a single strand of RNA or DNA, each polymer traverses the membrane as an extended chain that partially blocks the channel. The passage of each molecule is detected as a transient decrease of ionic current whose duration is proportional to polymer length. Channel blockades can therefore be used to measure polynucleotide length. With further improvements, the method could in principle provide direct, high-speed detection of the sequence of bases in single molecules of DNA or RNA.
Figures
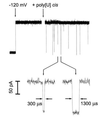
Figure 1
Oligomers of poly[U] caused transient blockades in the α-hemolysin single channel current. At the first arrow, a potential of −120 mV was applied across the membrane (cis side negative). This voltage caused a continuous current of −120 pA to flow. At the second arrow, poly[U] of mean length 210 bases was stirred into the cis compartment to a final concentration of 0.1 mg/ml. The polynucleotides caused short-lived current blockades. The_Inset_ (expanded time scale) shows two typical blockades with lifetimes of 300 and 1300 μs. For the purposes of illustration, the low time resolution current recordings (a total of 4 sec is shown here) were digitally filtered using an exponential smoothing moving average algorithm.
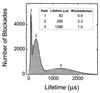
Figure 2
The characteristic lifetimes of channel blockades caused by poly[U] (0.1 mg/ml; mean length, 210 nt) fell within three well-defined peaks. The mean lifetimes corresponding to the three peaks were determined by fitting the sum of three Gaussians to the data.
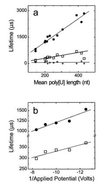
Figure 3
Poly[U]-induced channel blockade lifetimes were proportional to (a) mean polymer length and (b) inversely proportional to applied voltage. The plots show lifetimes for (a) peaks 1 (+), 2 (□), and 3 (•) in experiments using_V_ = −120 mV with 13 different size selected poly[U]s and (b) for peaks 2 (□), and 3 (•) with poly[U] of mean length 215 nt at the indicated voltages. Although the peak 1 lifetime appeared to be independent of the applied voltage (data not shown), because this lifetime (≈100 μs) was barely a factor of 2 greater than the time resolution of our system, a slight voltage dependence could not be ruled out.
Figure 4
Single-stranded DNA traversed the α-hemolysin pore. The PCR-amplified product (lane 1) of a 40 μl sample of the trans chamber compared with (lane 2) the similarly amplified product of a 109-fold diluted sample of the cis chamber. All samples were taken following an 8 channel experiment in which a mixture of 150-nt single-stranded DNA (see Materials and Methods) and 100-nt double-stranded DNA was added to the cis chamber. Material in the trans chamber sample reflects accumulation during application of −120 mV (cis chamber negative) for 42 min. Lanes 3-7: example of competitive PCR analysis (25) to show accumulation of single-stranded molecules in the trans chamber. The results show that the 40-μl sample used here contained ≈400 of the 150-nt single-stranded molecules. To each of five 40-μl samples of the trans chamber were mixed (lanes 3–7) 0, 100, 200, 400, or 800 molecules of a competing 120-nt DNA that was PCR amplified using the same primers as the 150-nt DNA. The position of the amplified 150-, 120-, and 100-nt polymers is shown at left. Lanes 1 and 3 show independently amplified 40-μl samples of the same trans chamber.
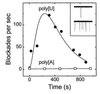
Figure 5
Enzymatic hydrolysis of poly[U] caused a transient increase in the rate of oligonucleotide-induced channel blockades. In two separate experiments, at time 0, ribonuclease A (bovine pancreatic RNase Type XII-A; Sigma or Worthington) was added to the cis chamber containing either 0.13 mg/ml poly[U] (•) (mean length, 1100 nt) or 0.27 mg/ml poly[A] (□) (mean length, 20,000 nt; Fluka). The final ribonuclease concentrations were 0.04 mg/ml (poly[U] experiment) or 0.13 mg/ml (poly[A] experiment). Blockade rates were calculated from single channel recordings. The Inset shows (null)/1;3-sec sample traces for the poly[U] experiment before (Upper) and ≈30 sec after (Lower) addition of ribonuclease. Similar experiments with a poly[U] sample of greater length (3000 nt) yielded results comparable to those obtained with the 1100-nt poly[U]. The current was digitized using a SONY/Dagan DAT recorder at 48,000 points/sec.
Similar articles
- Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules.
Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Akeson M, et al. Biophys J. 1999 Dec;77(6):3227-33. doi: 10.1016/S0006-3495(99)77153-5. Biophys J. 1999. PMID: 10585944 Free PMC article. - Driven DNA transport into an asymmetric nanometer-scale pore.
Henrickson SE, Misakian M, Robertson B, Kasianowicz JJ. Henrickson SE, et al. Phys Rev Lett. 2000 Oct 2;85(14):3057-60. doi: 10.1103/PhysRevLett.85.3057. Phys Rev Lett. 2000. PMID: 11006002 - Rapid nanopore discrimination between single polynucleotide molecules.
Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Meller A, et al. Proc Natl Acad Sci U S A. 2000 Feb 1;97(3):1079-84. doi: 10.1073/pnas.97.3.1079. Proc Natl Acad Sci U S A. 2000. PMID: 10655487 Free PMC article. - Multichannel simultaneous measurements of single-molecule translocation in alpha-hemolysin nanopore array.
Osaki T, Suzuki H, Le Pioufle B, Takeuchi S. Osaki T, et al. Anal Chem. 2009 Dec 15;81(24):9866-70. doi: 10.1021/ac901732z. Anal Chem. 2009. PMID: 20000639 - Nanopores and nucleic acids: prospects for ultrarapid sequencing.
Deamer DW, Akeson M. Deamer DW, et al. Trends Biotechnol. 2000 Apr;18(4):147-51. doi: 10.1016/s0167-7799(00)01426-8. Trends Biotechnol. 2000. PMID: 10740260 Review.
Cited by
- Electron beam-assisted healing of nanopores in magnesium alloys.
Zheng H, Liu Y, Cao F, Wu S, Jia S, Cao A, Zhao D, Wang J. Zheng H, et al. Sci Rep. 2013;3:1920. doi: 10.1038/srep01920. Sci Rep. 2013. PMID: 23719630 Free PMC article. - Crown ether-electrolyte interactions permit nanopore detection of individual DNA abasic sites in single molecules.
An N, Fleming AM, White HS, Burrows CJ. An N, et al. Proc Natl Acad Sci U S A. 2012 Jul 17;109(29):11504-9. doi: 10.1073/pnas.1201669109. Epub 2012 Jun 18. Proc Natl Acad Sci U S A. 2012. PMID: 22711805 Free PMC article. - Single-molecule sensing electrode embedded in-plane nanopore.
Tsutsui M, Rahong S, Iizumi Y, Okazaki T, Taniguchi M, Kawai T. Tsutsui M, et al. Sci Rep. 2011;1:46. doi: 10.1038/srep00046. Epub 2011 Jul 28. Sci Rep. 2011. PMID: 22355565 Free PMC article. - Single-cell RNA-sequencing: The future of genome biology is now.
Picelli S. Picelli S. RNA Biol. 2017 May 4;14(5):637-650. doi: 10.1080/15476286.2016.1201618. Epub 2016 Jul 21. RNA Biol. 2017. PMID: 27442339 Free PMC article. Review. - Dynamic interaction of fluoroquinolones with magnesium ions monitored using bacterial outer membrane nanopores.
Wang J, Prajapati JD, Kleinekathöfer U, Winterhalter M. Wang J, et al. Chem Sci. 2020 Aug 31;11(38):10344-10353. doi: 10.1039/d0sc03486j. Chem Sci. 2020. PMID: 34094296 Free PMC article.
References
- Neher E, Sakmann B. Nature (London) 1976;260:799–802. - PubMed
- Hille B. Ionic Channels of Excitable Membranes. 2nd Ed. Sunderland, MA: Sinauer; 1992.
- Sigworth F J. Q Rev Biophys. 1994;27:1–40. - PubMed
- Keynes R D. Q Rev Biophys. 1994;27:339–434. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources