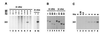In vitro V(D)J recombination: signal joint formation - PubMed (original) (raw)
In vitro V(D)J recombination: signal joint formation
P Cortes et al. Proc Natl Acad Sci U S A. 1996.
Abstract
The first step of V(D)J recombination, specific cleavage at the recombination signal sequence (RSS), can be carried out by the recombination activating proteins RAG1 and RAG2. In vivo, the cleaved coding and signal ends must be rejoined to generate functional antigen receptors and maintain chromosomal integrity. We have investigated signal joint formation using deletion and inversion substrates in a cell free system. RAG1 and RAG2 alone or in combination were unable to generate signal joints. However, RAG1 and RAG2 complemented with nuclear extracts were able to recombine an extrachromosomal substrate and form precise signal joints. The in vitro reaction resembled authentic V(D)J recombination in being Ku-antigen-dependent.
Figures
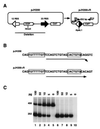
Figure 1
PCR assay for signal joint formation. (A) Schematic representation of the pJH200 substrate (28). The unrecombined plasmid and the recombined product are shown on the left and right, respectively. The relative positions of the primers (R5 and R14) used to amplify the 252-bp region of the recombined product are indicated by arrows (not to scale). (B) Nucleotide sequence to which primer R5 hybridizes in the unrecombined (pJH200) and recombined (pJH200+R) plasmids. Note that in the unrecombined substrate there is a 1-bp mismatch at the 3′ end of R5 (arrowhead). (C) Evaluation of primer combination R5/R14 on recombined and unrecombined sequences. The PCR assay was performed using the indicated amounts of recombined plasmid as template in the presence (lanes 1–5) or absence (lanes 6–10) of 2.5 ng of unrecombined substrate. The 456- and 252-bp fragments are the amplified products from pJH200 and pJH200+R, respectively.
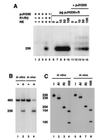
Figure 2
In vitro signal joint formation by deletion. (A) In vitro signal joint formation by deletion requires RAG1, RAG2, and nuclear extracts. pJH200 was incubated at 30°C with the indicated combinations of purified RAG1 (R1), RAG2 (R2), and nuclear extracts (NE) from 293-S cells under the conditions describe. The DNA was then purified and analyzed by PCR assay as described in Fig. 1. The autoradiograph from one such experiment is presented on the left (lanes 1–9), while the right (lanes 10 and 11) shows the 32P-labeled PCR products amplified from _in vivo_-recombined DNA purified from 293T cells transfected with pJH200 alone (−) or cotransfected with RAG1 and RAG2 (+). Lane 9 correspond to a control reaction, RAG1, RAG2, and NE but not template DNA. (B) Time course of in vitro signal joint formation. The reaction conditions were identical to those described in A, lane 7 (with RAGs and nuclear extract) and the products were analyzed as explained in Fig. 1. Lane 11 is identical to lane 10 except that template DNA was not included in the reaction. (C) Digestion pattern of the_in vivo_- and _in vitro_-recombined products. Purified 252-bp 32P-labeled PCR products were incubated with no enzyme (−), _Rsa_I (RI), or_Hin_dIII (HIII) and separated on an 8% polyacrylamide/1× TBE gel. The 252-bp fragment contains a unique_Hin_dIII site 151 bp from the R14 oligonucleotide. This fragment also contains two _Rsa_I sites, one in the 23 RSS and the other 25 bp from oligonucleotide R14. (D) Different nuclear extracts can complement RAG proteins in signal joint formation. RAG1 (R1) and RAG2 (R2) were supplemented with extracts prepared from BASC6C2 (BASC), 22D6, HeLa, CHO-S, or 293-S cells, as indicated. Each sample was processed as in A and the bands were visualized by autoradiography.
Figure 3
Signal joints generated in vitro and detected by PCR are precise. (A) Southern blot analysis of pJH200 recombined in vitro and PCR-amplified with flanking oligonucleotides R3 and R14. Because primers R3 and R14 hybridize equally well to the recombined and unrecombined pJH200, the DNA recovered from in vitro recombination reactions was first digested with _Hin_cII before PCR amplification, to enrich for the recombined DNA. The in vitro recombination assay was performed with the indicated combinations of RAGs, 293-S nuclear extracts, and 50 ng of pJH200 (lanes 1–5). The_Hin_cII-digested DNA from these recombination reactions, as well as a titration (0–100 pg) of pJH200+R without (lanes 6–10) or with (lanes 11–15) 2.5 ng of pJH200, was then subjected to PCR with the following modifications: no [α32P]dCTP was added to the mixture and the primer pair used was R3/R14 (which generates a 256-bp fragment). Unlike R5, which overlaps the signal joint, the 3′ end of R3 stops 3 bp before the signal joint and is, therefore, considered a flanking primer. The PCR products were separated on an agarose gel, transferred to nylon membrane, subjected to Southern blot analysis using a 32P-labeled oligonucleotide that overlaps the signal joint and visualized by autoradiography. (B) pJH200 recombined in vitro and PCR-amplified with flanking oligonucleotides R3 and R14, in the presence of [32P]dCTP. pJH200 was subjected to the in vitro recombination assay in the absence (lane 1) or presence of RAGs and 293-S nuclear extract (lane 2). As a control, pJH200 was transiently transfected into 293T cells alone (lane 3) or along with RAGs (lane 4) to generate in vivo_-recombined plasmid. DNA recovered from recombination reactions was digested with_Hin_cII and PCR-amplified using flanking oligonucleotides R3 and R14. [α32P]dCTP was used during PCR amplification. The 32P-labeled PCR products were fractionated on an 8% native polyacrylamide/1× TBE gel and detected by autoradiography. (C) Restriction enzyme digestion of signal joints formed in vivo and in vitro. The 256-bp product from in vivo and_in vitro reactions was extracted from a gel and submitted to restriction enzyme digestion with _Apa_L1 (A1), _Rsa_I (RI), and _Hin_dIII (HIII). Undigested and digested aliquots were separated on an 12% native polyacrylamide gel and the 32P-labeled PCR products were detected by autoradiography. _Apa_L1 cuts at the correct signal joint, located 20 bp from the end of the PCR product.
Figure 4
In vitro signal joint formation by inversion. (A) Autoradiogram of the 32P-labeled PCR products amplified from the recombination reaction using oligonucleotides R5 and R14 and separated in an 8% polyacrylamide/1× TBE gel. pJH288 inversion substrate was incubated with RAG1 (R1), RAG2 (R2), and nuclear extract (NE) as indicated. The_in vivo_ results, to the right, show the32P-labeled PCR products from 293T cells transfected with pJH288 alone (−) or cotransfected with RAG1 and RAG2 (+). The 231-bp32P-labeled PCR product is indicated. (B) Digestion pattern of the in vivo and in vitro products, recombined by inversion. The 231-bp32P-labeled PCR products were incubated with no enzyme (−), _Apa_LI (A1), _Bam_HI (BI), and_Sma_I (SI) as indicated and separated in an 8% polyacrylamide/1× TBE gel. The sizes of the fragments are indicated. (C) Time course of in vitro signal joint formation by invertion. The reaction conditions were identical to those described in A, lane 7 (with RAGs and nuclear extract).
Figure 5
Anti- (α) Ku70 antibodies specifically inhibit_in vitro_ signal joint formation. On the left (lanes 1–5), the in vitro recombination assay was performed with the indicated combinations of RAGs, 293-S nuclear extracts, and 50 ng of pJH200. Lane 4 corresponds to a control reaction: RAG1, RAG2, and NE but no template DNA. Lanes 6–11 show complete reactions in which the RAGs and nuclear extracts were preincubated for 15 min at 4°C with a preimmune sera (PI), a polyclonal αKu70 (I-αKu70) raised against rat Ku-70, αKu monoclonal (m-αKu70/80) raised to the human p70/p86 heterodimer (ref. , monoclonal 162), or an isotype-matched monoclonal antibody (HB95; American Type Culture Collection). The 1× and 2× refer to the relative amounts of sera used. Each in vitro recombination reaction was then analyzed by PCR as described in Fig. 1.
Similar articles
- Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends.
Jones JM, Gellert M. Jones JM, et al. Proc Natl Acad Sci U S A. 2001 Nov 6;98(23):12926-31. doi: 10.1073/pnas.221471198. Epub 2001 Oct 23. Proc Natl Acad Sci U S A. 2001. PMID: 11606753 Free PMC article. - Ku86 is not required for protection of signal ends or for formation of nonstandard V(D)J recombination products.
Han JO, Steen SB, Roth DB. Han JO, et al. Mol Cell Biol. 1997 Apr;17(4):2226-34. doi: 10.1128/MCB.17.4.2226. Mol Cell Biol. 1997. PMID: 9121473 Free PMC article. - RAG1 and RAG2 in V(D)J recombination and transposition.
Fugmann SD. Fugmann SD. Immunol Res. 2001;23(1):23-39. doi: 10.1385/IR:23:1:23. Immunol Res. 2001. PMID: 11417858 Review. - Analysis of variable (diversity) joining recombination in DNAdependent protein kinase (DNA-PK)-deficient mice reveals DNA-PK-independent pathways for both signal and coding joint formation.
Bogue MA, Jhappan C, Roth DB. Bogue MA, et al. Proc Natl Acad Sci U S A. 1998 Dec 22;95(26):15559-64. doi: 10.1073/pnas.95.26.15559. Proc Natl Acad Sci U S A. 1998. PMID: 9861008 Free PMC article. - V(D)J recombination: double-strand break repair gene products used in the joining mechanism.
Weaver D, Boubnov N, Wills Z, Hall K, Staunton J. Weaver D, et al. Ann N Y Acad Sci. 1995 Sep 29;764:99-111. doi: 10.1111/j.1749-6632.1995.tb55811.x. Ann N Y Acad Sci. 1995. PMID: 7486596 Review. No abstract available.
Cited by
- Metabolic sensor AMPK directly phosphorylates RAG1 protein and regulates V(D)J recombination.
Um JH, Brown AL, Singh SK, Chen Y, Gucek M, Lee BS, Luckey MA, Kim MK, Park JH, Sleckman BP, Gellert M, Chung JH. Um JH, et al. Proc Natl Acad Sci U S A. 2013 Jun 11;110(24):9873-8. doi: 10.1073/pnas.1307928110. Epub 2013 May 28. Proc Natl Acad Sci U S A. 2013. PMID: 23716691 Free PMC article. - V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins.
Sawchuk DJ, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenzweig MC, Cortes P. Sawchuk DJ, et al. J Exp Med. 1997 Jun 2;185(11):2025-32. doi: 10.1084/jem.185.11.2025. J Exp Med. 1997. PMID: 9166431 Free PMC article. - SARS-CoV envelope protein palmitoylation or nucleocapid association is not required for promoting virus-like particle production.
Tseng YT, Wang SM, Huang KJ, Wang CT. Tseng YT, et al. J Biomed Sci. 2014 Apr 27;21(1):34. doi: 10.1186/1423-0127-21-34. J Biomed Sci. 2014. PMID: 24766657 Free PMC article. - V(D)J recombination: in vitro coding joint formation.
Weis-Garcia F, Besmer E, Sawchuk DJ, Yu W, Hu Y, Cassard S, Nussenzweig MC, Cortes P. Weis-Garcia F, et al. Mol Cell Biol. 1997 Nov;17(11):6379-85. doi: 10.1128/MCB.17.11.6379. Mol Cell Biol. 1997. PMID: 9343399 Free PMC article.
References
- Lewis S. Adv Immunol. 1994;56:27–151. - PubMed
- Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Science. 1993;261:1175–1178. - PubMed
- Komori T, Okada A, Stewart V, Alt F W. Science. 1993;261:1171–1175. - PubMed
- Schatz D, Baltimore D. Cell. 1988;53:107–115. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous