The AML1/ETO fusion protein activates transcription of BCL-2 - PubMed (original) (raw)
The AML1/ETO fusion protein activates transcription of BCL-2
L Klampfer et al. Proc Natl Acad Sci U S A. 1996.
Abstract
The AML1 gene, located on chromosome 21, is involved in several distinct chromosomal translocations in human leukemia. In t(8;21) acute myelogenous leukemia (AML), the AML1 gene is juxtaposed to the ETO gene located on chromosome 8, generating an AML1/ETO fusion protein. Both AML1/ETO and the AML1 proteins recognize the same consensus DNA-binding motif (TGT/CGGT), which is found in the promoters of several genes involved in hematopoiesis. We found that two myeloid leukemia cell lines with the t(8;21) translocation, Kasumi and SKNO-1, have elevated levels of BCL-2 protein compared with other myeloid cell lines. In addition, we identified a consensus AML1 binding site in the BCL-2 promoter. Thus far, AML1/ETO has been shown to dominantly repress its target genes; however, we found that AML1/ETO activates transcription of the BCL-2 gene in U937 cells. This activation requires the presence of both the runt homology domain (rhd) and the C-terminal portion of AML1/ETO. We demonstrated sequence specific binding of both AML1A and AML1/ETO to the TGTGGT sequence in the BCL-2 promoter and showed that the AML1 binding site is required for responsiveness to AML1/ETO. Interestingly, AML1A and AML1B do not modulate the activity of the BCL-2 promoter. The elevated levels of BCL-2 in cells that express AML1/ETO may prolong their life span and contribute to the development of t(8;21) leukemia.
Figures
Figure 2
Schematic representation of the BCL-2 5′untranslated region. The location of the P1 and P2 promoters,_Sac_I fragment, and the potential AML1 binding site (shown in bold and underlined) are indicated. The sequence shown is contained in the wild-type oligonucleotide used for the EMSA.
Figure 1
Expression of BCL-2, MCL-1 and BCL-x proteins in myeloid leukemia cell lines. (A) Whole cell lysates from Kasumi, U937, KG-1, K562, TF-1 and SKNO-1 cells were separated on a 12% SDS/PAGE. The gel was stained with Coomassie blue after transfer to demonstrate equal loading of proteins (not shown). The levels of BCL-2 were detected by Western blot analysis, using antibodies specific for BCL-2 proteins. The membrane was stripped of bound antibody and reprobed with an antibody specific for MCL-1. (B) The cell lysates used in Fig. 1_A_ were tested for the presence of the BCL-x protein by Western blot analysis, using antibodies that specifically recognize BCL-x protein.
Figure 3
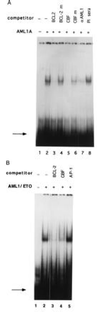
AML1A and AML1/ETO bind to the BCL-2 promoter. (A) A 32P-labeled oligonucleotide containing the AML1 binding site from the BCL-2 promoter was run alone (lane 1), or incubated with 5 μl of bacterially expressed AML1A protein (lane 2). The reaction mixture was preincubated with a 100-fold molar excess of unlabeled wild-type oligonucleotide (lane 3) or an oligonucleotide with a mutated AML1-binding site (lane 4), an oligonucleotide containing the AML1 site from the IL-3 promoter (lane 5) or the IL-3 promoter oligonucleotide with a mutated AML1 site (lane 6). In lane 7, the AML1A protein was preincubated with 2 μl of AML1 antiserum, and in lane 8 with 2 μl of preimmune serum. The binding of AML1A to the BCL-2 promoter was analyzed by EMSA. The position of the free probe is indicated by the arrow. (B) The 32P-labeled 40-mer oligonucleotide (shown in Fig. 2) was incubated with 5 μl of bacterially expressed AML1/ETO alone (lane 2), or with a 100-fold molar excess of unlabeled BCL-2 oligonucleotide (lane 3), a 100-fold excess of unlabeled oligonucleotide containing the AML1 binding site from the IL-3 promoter (lane 4) or a 100-fold excess of unlabeled oligonucleotide containing a consensus AP-1 site (lane 5). No protein was added in lane 1. The position of the free probe is indicated by the arrow.
Figure 4
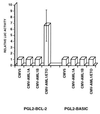
AML1/ETO transactivates the BCL-2 promoter. U937 cells were cotransfected with pGL2-basic or pGL2-BCL-2 reporter gene constructs and with pCMV-AML1A, pCMV-AML1B or pCMV-AML1/ETO expression vectors (together with pGHX5 to normalize transfection efficiency). Promoter activity was measured as the ratio of the luciferase activity and the concentration of growth hormone in the conditioned media. The promoter activity in the presence of the empty CMV5 plasmid was defined as 1 and the promoter activity in the presence of expression vectors for AML1A, AML1B, and AML1/ETO was defined relative to that value. The results shown are the mean of four different experiments (SEs are shown when measurable).
Figure 5
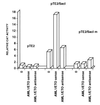
The AML1 binding site in the _Sac_I fragment confers AML1/ETO responsiveness to a heterologous promoter. U937 cells were cotransfected with the pTE2, pTE2/_Sac_I or pTE2/_Sac_Im plasmids and with AML1/ETO expression vector (AML1/ETO sense), with its antisense derivative (AML1/ETO antisense) or with an empty pCIneo vector (0). The activity of the pTE2 plasmid in the presence of an empty vector was assigned a value of one. Results from one of three independent experiments are shown.
Figure 6
(A) Schematic representation and transcriptional activity of the various AML1/ETO deletion mutants. RHD, rhd; TAF, TATA-binding-protein-associated factor-110 homology domain; PST, proline-serine-threonine-rich region; ZF, zinc finger domain. (B) Transcriptional activity of the wild-type AML1/ETO and mutant AML/ETO proteins. U937 cells were cotransfected with the pGL2-BCL2 plasmid and the wild-type CMV5-AML1/ETO expression plasmid or with its deletion mutants as shown in Fig. 6_A_. Luciferase activity in the presence of the CMV5 plasmid was assigned a value of 1. Results represent the mean of four independent experiments.
Similar articles
- The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B.
Frank R, Zhang J, Uchida H, Meyers S, Hiebert SW, Nimer SD. Frank R, et al. Oncogene. 1995 Dec 21;11(12):2667-74. Oncogene. 1995. PMID: 8545124 - The t(8;21) fusion protein, AML1/ETO, transforms NIH3T3 cells and activates AP-1.
Frank RC, Sun X, Berguido FJ, Jakubowiak A, Nimer SD. Frank RC, et al. Oncogene. 1999 Mar 4;18(9):1701-10. doi: 10.1038/sj.onc.1202459. Oncogene. 1999. PMID: 10208431 - Synergistic up-regulation of the myeloid-specific promoter for the macrophage colony-stimulating factor receptor by AML1 and the t(8;21) fusion protein may contribute to leukemogenesis.
Rhoades KL, Hetherington CJ, Rowley JD, Hiebert SW, Nucifora G, Tenen DG, Zhang DE. Rhoades KL, et al. Proc Natl Acad Sci U S A. 1996 Oct 15;93(21):11895-900. doi: 10.1073/pnas.93.21.11895. Proc Natl Acad Sci U S A. 1996. PMID: 8876234 Free PMC article. - The AML1 and ETO genes in acute myeloid leukemia with a t(8;21).
Nucifora G, Rowley JD. Nucifora G, et al. Leuk Lymphoma. 1994 Aug;14(5-6):353-62. doi: 10.3109/10428199409049690. Leuk Lymphoma. 1994. PMID: 7812194 Review. - The AML1 gene: a transcription factor involved in the pathogenesis of myeloid and lymphoid leukemias.
Lo Coco F, Pisegna S, Diverio D. Lo Coco F, et al. Haematologica. 1997 May-Jun;82(3):364-70. Haematologica. 1997. PMID: 9234595 Review.
Cited by
- MicroRNA let-7b downregulates AML1-ETO oncogene expression in t(8;21) AML by targeting its 3'UTR.
Johnson DT, Davis AG, Zhou JH, Ball ED, Zhang DE. Johnson DT, et al. Exp Hematol Oncol. 2021 Feb 2;10(1):8. doi: 10.1186/s40164-021-00204-7. Exp Hematol Oncol. 2021. PMID: 33531067 Free PMC article. - TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia.
Sabaawy HE, Azuma M, Embree LJ, Tsai HJ, Starost MF, Hickstein DD. Sabaawy HE, et al. Proc Natl Acad Sci U S A. 2006 Oct 10;103(41):15166-71. doi: 10.1073/pnas.0603349103. Epub 2006 Oct 2. Proc Natl Acad Sci U S A. 2006. PMID: 17015828 Free PMC article. - AML1/ETO oncoprotein is directed to AML1 binding regions and co-localizes with AML1 and HEB on its targets.
Gardini A, Cesaroni M, Luzi L, Okumura AJ, Biggs JR, Minardi SP, Venturini E, Zhang DE, Pelicci PG, Alcalay M. Gardini A, et al. PLoS Genet. 2008 Nov;4(11):e1000275. doi: 10.1371/journal.pgen.1000275. Epub 2008 Nov 28. PLoS Genet. 2008. PMID: 19043539 Free PMC article. - Identification and characterization of novel AML1-ETO fusion transcripts in pediatric t(8;21) acute myeloid leukemia: a report from the Children's Oncology Group.
LaFiura KM, Edwards H, Taub JW, Matherly LH, Fontana JA, Mohamed AN, Ravindranath Y, Ge Y; Children's Oncology Group. LaFiura KM, et al. Oncogene. 2008 Aug 21;27(36):4933-42. doi: 10.1038/onc.2008.134. Epub 2008 May 12. Oncogene. 2008. PMID: 18469864 Free PMC article. - Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO.
Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, Lazar MA. Gelmetti V, et al. Mol Cell Biol. 1998 Dec;18(12):7185-91. doi: 10.1128/MCB.18.12.7185. Mol Cell Biol. 1998. PMID: 9819405 Free PMC article.
References
- Nichols J, Nimer S D. Blood. 1992;80:2953–2963. - PubMed
- Erickson P, Gao K S, Chang T, Look T, Whisenant E, Raimondi S, Lasher J, Trujillo J, Rowley J, Drabkin H. Blood. 1992;80:1825–1831. - PubMed
- Frank R, Zhang J, Hiebert S W, Meyers S, Nimer S D. Oncogene. 1995;11:2667–2674. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources