A conserved repetitive DNA element located in the centromeres of cereal chromosomes - PubMed (original) (raw)
Comparative Study
A conserved repetitive DNA element located in the centromeres of cereal chromosomes
J Jiang et al. Proc Natl Acad Sci U S A. 1996.
Abstract
Repetitive DNA sequences have been demonstrated to play an important role for centromere function of eukaryotic chromosomes, including those from fission yeast, Drosophila melanogaster, and humans. Here we report on the isolation of a repetitive DNA element located in the centromeric regions of cereal chromosomes. A 745-bp repetitive DNA clone pSau3A9, was isolated from sorghum (Sorghum bicolor). This DNA element is located in the centromeric regions of all sorghum chromosomes, as demonstrated by fluorescence in situ hybridization. Repetitive DNA sequences homologous to pSau3A9 also are present in the centromeric regions of chromosomes from other cereal species, including rice, maize, wheat, barley, rye, and oats. Probe pSau3A9 also hybridized to the centromeric region of B chromosomes from rye and maize. The repetitive nature and its conservation in distantly related plant species indicate that the pSau3A9 family may be associated with centromere function of cereal chromosomes. The absence of DNA sequences homologous to pSau3A9 in dicot species suggests a faster divergence of centromererelated sequences compared with the telomere-related sequences in plants.
Figures
Figure 1
DNA of sorghum BAC clone 52A4 was digested with restriction enzymes _Dra_I (lane 1), _Sal_I (lane 2), _Sau_3AI (lane 3), _Acc_I (lane 4), and _Hin_dIII (lane 5). The same blot was probed with genomic DNA from wheat, maize, and rice, respectively. A 745_Sau_3AI fragment (arrow) hybridized to the genomic DNA from all the three species. This fragment was subcloned as plasmid pSau3A9.
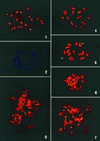
Figure 2
FISH analysis using pSau3A9 as a probe: 1, sorghum; 2, rice; 3, wheat; 4, barley; 5, rye, 14 A and 2 B (arrows) chromosomes have similar FISH signals at centromeres; 6, maize; 7, oat.
Figure 3
Genomic DNA from sorghum (lane 1), maize (lane 2), wheat (lane 3), rice (lane 4), A. thaliana (lane 5), tobacco (lane 6), tomato (lane 7), and soybean (lane 8) was digested with _Sal_I and probed with pSau3A9. Strong hybridization signals were detected in the four cereal species, and very weak or no signals were detected in the four dicot species.
Figure 4
Nucleotide sequence of pSau3A9.
Similar articles
- A Ty3/gypsy retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes.
Presting GG, Malysheva L, Fuchs J, Schubert I. Presting GG, et al. Plant J. 1998 Dec;16(6):721-8. doi: 10.1046/j.1365-313x.1998.00341.x. Plant J. 1998. PMID: 10069078 - A cereal centromeric sequence.
Aragón-Alcaide L, Miller T, Schwarzacher T, Reader S, Moore G. Aragón-Alcaide L, et al. Chromosoma. 1996 Dec;105(5):261-8. doi: 10.1007/BF02524643. Chromosoma. 1996. PMID: 8939818 - Organization of the 1.9-kb repeat unit RCE1 in the centromeric region of rice chromosomes.
Nonomura KI, Kurata N. Nonomura KI, et al. Mol Gen Genet. 1999 Feb;261(1):1-10. doi: 10.1007/s004380050935. Mol Gen Genet. 1999. PMID: 10071204 - Centromere structure and function in budding and fission yeasts.
Carbon J, Clarke L. Carbon J, et al. New Biol. 1990 Jan;2(1):10-9. New Biol. 1990. PMID: 2078550 Review. - [Advances in research of the structure and function of plant centromeres].
She CW, Song YC. She CW, et al. Yi Chuan. 2006 Dec;28(12):1597-606. doi: 10.1360/yc-006-1597. Yi Chuan. 2006. PMID: 17138549 Review. Chinese.
Cited by
- Centromeres cluster de novo at the beginning of meiosis in Brachypodium distachyon.
Wen R, Moore G, Shaw PJ. Wen R, et al. PLoS One. 2012;7(9):e44681. doi: 10.1371/journal.pone.0044681. Epub 2012 Sep 7. PLoS One. 2012. PMID: 22970287 Free PMC article. - De Novo Centromere Formation and Centromeric Sequence Expansion in Wheat and its Wide Hybrids.
Guo X, Su H, Shi Q, Fu S, Wang J, Zhang X, Hu Z, Han F. Guo X, et al. PLoS Genet. 2016 Apr 25;12(4):e1005997. doi: 10.1371/journal.pgen.1005997. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27110907 Free PMC article. - Structure and genomic organization of centromeric repeats in Arabidopsis species.
Kawabe A, Nasuda S. Kawabe A, et al. Mol Genet Genomics. 2005 Feb;272(6):593-602. doi: 10.1007/s00438-004-1081-x. Epub 2004 Dec 7. Mol Genet Genomics. 2005. PMID: 15586291 - Chromodomains direct integration of retrotransposons to heterochromatin.
Gao X, Hou Y, Ebina H, Levin HL, Voytas DF. Gao X, et al. Genome Res. 2008 Mar;18(3):359-69. doi: 10.1101/gr.7146408. Epub 2008 Feb 6. Genome Res. 2008. PMID: 18256242 Free PMC article. - Molecular anatomy of a small chromosome in the green alga Chlorella vulgaris.
Noutoshi Y, Ito Y, Kanetani S, Fujie M, Usami S, Yamada T. Noutoshi Y, et al. Nucleic Acids Res. 1998 Sep 1;26(17):3900-7. doi: 10.1093/nar/26.17.3900. Nucleic Acids Res. 1998. PMID: 9705496 Free PMC article.
References
- Clarke L. Trends Genet. 1990;6:150–154. - PubMed
- Haaf T, Warburton P E, Willard H F. Cell. 1992;70:681–696. - PubMed
- Tyler-Smith C, Oakey R, Larin Z, Fisher R B, Crocker M, Affara N A, Ferguson-Smith M A, Muenke M, Zuffardi O, Jobling M A. Nat Genet. 1993;5:368–375. - PubMed
- Larin Z, Fricker M D, Tyler-Smith C. Hum Mol Genet. 1994;3:689–695. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources