A possible mitochondrial gene in the early-branching amitochondriate protist Trichomonas vaginalis - PubMed (original) (raw)
A possible mitochondrial gene in the early-branching amitochondriate protist Trichomonas vaginalis
A J Roger et al. Proc Natl Acad Sci U S A. 1996.
Abstract
Trichomonads are anaerobic flagellated protists that, based on analyses of ribosomal RNA sequences, represent one of the earliest branching lineages among the eukaryotes. The absence of mitochondria in these organisms coupled with their deep phylogenetic position has prompted several authors to suggest that trichomonads, along with other deeply-branching amitochondriate protist groups, diverged from the main eukaryotic lineage prior to the endosymbiotic origin of mitochondria. In this report we describe the presence of a gene in Trichomonas vaginalis specifically related to mitochondrial chaperonin 60 (cpn60). A recent study indicates that a protein immunologically related to cpn60 is located in trichomonad hydrogenosomes. Together, these data provide evidence that ancestors of trichomonads perhaps harbored the endosymbiotic progenitors of mitochondria, but that these evolved into hydrogenosomes early in trichomonad evolution.
Figures
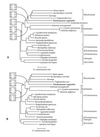
Figure 1
Phylogenies of cpn60 homologs. Sequences were selected from the database for organisms previously shown to branch in the region of the mitochondrial cpn60 clade (11, 12). The trees shown are derived from neighbor-joining analysis of a accepted point mutation corrected distance matrix. Percentage bootstrap support is shown above selected branches in boxes, from bootstrap analyses employing the protein maximum likelihood (ML), neighbor-joining distance (NJ), and maximum parsimony (MP) methods. (A) Cpn60 tree derived from the full alignment. The maximum likelihood tree (ln likelihood = −8933.6) differed from the neighbor-joining tree by the placement of the T. vaginalis and E. histolytica sequences as sister groups. Parsimony generated five trees of length = 2369 all of which differed principally from the tree shown by the placement of T. vaginalis as a sister group of the Rickettsiales species. Asterisks (∗) indicate that the method used did not recover this node in the majority of bootstrap replicates. (B) Cpn60 tree with the E. histolytica sequence excluded. Maximum likelihood yielded a tree of identical topology (ln likelihood = −12181.7), while parsimony generated three trees of length = 2209. Two of these differed from the neighbor-joining tree by the placement of the α-Protebacteria (excluding the Rickettsiales) as a sister group to the γ- and β-Proteobacteria. The third differed by placing T. vaginalis as an immediate relative to the Rickettsiales (see_Results_).
Figure 2
The impact of the sampling of Rickettsiales species on the bootstrap support for two alternative topologies of the cpn60 tree. The dataset excluding the E. histolytica sequence, was used to examine the bootstrap support for two alternative clades each indicated by • on the two trees. (A) The T. vaginalis/mitochondria clade found by neighbor-joining, maximum likelihood and two of the three maximum parsimony trees. (B) The T. vaginalis/Rickettsiales clade displayed by the third maximum parsimony tree (see Materials and Methods). Percentage bootstrap support for each clade is indicated to the left of the trees. Three different combinations of Rickettsiales species were used in the dataset. Species abbreviations are: Ec, Ehrlichia chaffeensis; Cr, C. ruminantium and Rt;R. tsutsugamushi. For each combination of species, bootstrap support for the clade was evaluated using the methods NJ (neighbor-joining distance), MP (maximum parsimony) and ML (protein maximum likelihood) as described in the Materials and Methods.
Similar articles
- Presence of a mitochondrial-type 70-kDa heat shock protein in Trichomonas vaginalis suggests a very early mitochondrial endosymbiosis in eukaryotes.
Germot A, Philippe H, Le Guyader H. Germot A, et al. Proc Natl Acad Sci U S A. 1996 Dec 10;93(25):14614-7. doi: 10.1073/pnas.93.25.14614. Proc Natl Acad Sci U S A. 1996. PMID: 8962101 Free PMC article. - A mitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria.
Roger AJ, Svärd SG, Tovar J, Clark CG, Smith MW, Gillin FD, Sogin ML. Roger AJ, et al. Proc Natl Acad Sci U S A. 1998 Jan 6;95(1):229-34. doi: 10.1073/pnas.95.1.229. Proc Natl Acad Sci U S A. 1998. PMID: 9419358 Free PMC article. - Phylogenetic relationships of class II fumarase genes from trichomonad species.
Gerbod D, Edgcomb VP, Noël C, Vanácová S, Wintjens R, Tachezy J, Sogin ML, Viscogliosi E. Gerbod D, et al. Mol Biol Evol. 2001 Aug;18(8):1574-84. doi: 10.1093/oxfordjournals.molbev.a003944. Mol Biol Evol. 2001. PMID: 11470849 - Molecular biology of the amitochondriate parasites, Giardia intestinalis, Entamoeba histolytica and Trichomonas vaginalis.
Vanacova S, Liston DR, Tachezy J, Johnson PJ. Vanacova S, et al. Int J Parasitol. 2003 Mar;33(3):235-55. doi: 10.1016/s0020-7519(02)00267-9. Int J Parasitol. 2003. PMID: 12670510 Review. - Early branching eukaryotes?
Embley TM, Hirt RP. Embley TM, et al. Curr Opin Genet Dev. 1998 Dec;8(6):624-9. doi: 10.1016/s0959-437x(98)80029-4. Curr Opin Genet Dev. 1998. PMID: 9914207 Review.
Cited by
- Proteins of the glycine decarboxylase complex in the hydrogenosome of Trichomonas vaginalis.
Mukherjee M, Brown MT, McArthur AG, Johnson PJ. Mukherjee M, et al. Eukaryot Cell. 2006 Dec;5(12):2062-71. doi: 10.1128/EC.00205-06. Eukaryot Cell. 2006. PMID: 17158739 Free PMC article. - Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes.
Gupta RS. Gupta RS. Microbiol Mol Biol Rev. 1998 Dec;62(4):1435-91. doi: 10.1128/MMBR.62.4.1435-1491.1998. Microbiol Mol Biol Rev. 1998. PMID: 9841678 Free PMC article. Review. - Prokaryotic and eukaryotic features observed on the secondary structures of Giardia SSU rRNAs and its phylogenetic implications.
Hwang UW. Hwang UW. Parasitol Res. 2007 Apr;100(5):1159-63. doi: 10.1007/s00436-007-0471-5. Epub 2007 Feb 6. Parasitol Res. 2007. PMID: 17279392 - Why metronidazole is active against both bacteria and parasites.
Samuelson J. Samuelson J. Antimicrob Agents Chemother. 1999 Jul;43(7):1533-41. doi: 10.1128/AAC.43.7.1533. Antimicrob Agents Chemother. 1999. PMID: 10390199 Free PMC article. Review. No abstract available. - Diversity and origins of anaerobic metabolism in mitochondria and related organelles.
Stairs CW, Leger MM, Roger AJ. Stairs CW, et al. Philos Trans R Soc Lond B Biol Sci. 2015 Sep 26;370(1678):20140326. doi: 10.1098/rstb.2014.0326. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26323757 Free PMC article. Review.
References
- Gunderson J, Hinkle G, Leipe D L, Morrison H G, Stickel S K, Odelson D A, Breznak J A, Nerad T A, Müller M, Sogin M L. J Eukaryotic Microbiol. 1995;42:411–415. - PubMed
- Sogin M L. Curr Opin Genet Dev. 1991;1:457–463. - PubMed
- Patterson D J, Sogin M L. In: The Origin and Evolution of Prokaryotic and Eukaryotic Cells. Hartman H, Matsuno K, editors. Singapore: World Scientific; 1992. pp. 13–46.
- Müller M. J Gen Microbiol. 1993;139:2879–2889. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials