The mutation rate and cancer - PubMed (original) (raw)
The mutation rate and cancer
I P Tomlinson et al. Proc Natl Acad Sci U S A. 1996.
Abstract
The selection of advantageous mutations underlies tumorigenesis. The growth of a tumor is therefore a form of evolution at the somatic level, in which the population is comprised of individual cells within the tumor. Models of tumorigenesis have considered the relative importance of mutation and selection. We show that selection is more important than an increased mutation rate in the growth of a tumor. Some cancers may acquire a "mutator phenotype," probably leading to faster growth, but mutator phenotypes are not necessary for carcinogenesis.
Figures
Figure 1
Genetic model of colorectal tumorigenesis. This model illustrates the stepwise nature of tumorigenesis and the action of recessive (tumor-suppressor) and dominant (oncogene) mutations.
Figure 2
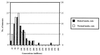
Model of tumorigenesis with two mutations required for tumor growth. The premises of the model are as follows. Given enough time, any stem cell will acquire the necessary mutations to become a tumor. In practice, organisms die before this occurs in nearly all of their stem cells. If, however, a stem cell acquires the necessary mutations exceptionally rapidly—whether by chance, after exposure to extrinsic mutagens, or as a result of an inherited tendency to cancer—a tumor will start to grow within an individual’s normal lifespan. We have taken a single stem cell and assigned to it probabilities of mutation per generation at tumor-suppressor and DNA repair loci. Here, the normal mutation rate is assumed to be 10−8 and the raised rate, 10−4 (see text). A simulation is then run to determine the time it takes for one cell to acquire the mutations needed to start tumor growth. (The computer model is available from the authors.) This simulation has been repeated for a total of 100 stem cells. [An otherwise identical model assuming a normal mutation rate of 10−6 (and thus taking far less computing time) has been run for 10,000 cells and confirms the results of this model.] The model is not quantitative (because we have not considered the many thousands of stem cells that probably exist in the human intestine, for example). Despite this, we can reasonably argue that the tumors which start to grow earliest will be those that occur most often in reality. The graph shows the number of tumors starting to grow with a raised (solid bars) and normal (open bars) mutation rate (y axis) and the cell generation number at which the tumor starts to grow (x axis; each bar representing divisions of 50 million generations, with bar 1 corresponding to 0–50 million generations, bar 2 to 51–100 million generations, etc.). While an equal number of tumors grow, in theory, with normal and raised mutation rates, the tumors that grow earliest (and therefore those that will occur in reality) usually start to grow with a normal mutation rate in this model.
Figure 3
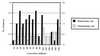
Model of tumorigenesis in an HNPCC patient with two mutations required for tumor growth. The model is as in Fig. 2, but the cell already possesses one mutation at the DNA repair locus. Two repair mutations raise the mutation rate by only 101. A significant proportion of the earliest tumors starts to grow with a normal mutation rate.
Similar articles
- Efficiency of carcinogenesis with and without a mutator mutation.
Beckman RA, Loeb LA. Beckman RA, et al. Proc Natl Acad Sci U S A. 2006 Sep 19;103(38):14140-5. doi: 10.1073/pnas.0606271103. Epub 2006 Sep 11. Proc Natl Acad Sci U S A. 2006. PMID: 16966602 Free PMC article. - Mutator mutations enhance tumorigenic efficiency across fitness landscapes.
Beckman RA. Beckman RA. PLoS One. 2009 Jun 10;4(6):e5860. doi: 10.1371/journal.pone.0005860. PLoS One. 2009. PMID: 19517009 Free PMC article. - Negative clonal selection in tumor evolution.
Beckman RA, Loeb LA. Beckman RA, et al. Genetics. 2005 Dec;171(4):2123-31. doi: 10.1534/genetics.105.040840. Epub 2005 Sep 2. Genetics. 2005. PMID: 16143627 Free PMC article. - Cancers exhibit a mutator phenotype: clinical implications.
Loeb LA, Bielas JH, Beckman RA. Loeb LA, et al. Cancer Res. 2008 May 15;68(10):3551-7; discussion 3557. doi: 10.1158/0008-5472.CAN-07-5835. Cancer Res. 2008. PMID: 18483233 Review. - Efficiency of carcinogenesis: is the mutator phenotype inevitable?
Beckman RA. Beckman RA. Semin Cancer Biol. 2010 Oct;20(5):340-52. doi: 10.1016/j.semcancer.2010.10.004. Epub 2010 Oct 8. Semin Cancer Biol. 2010. PMID: 20934514 Review.
Cited by
- Pretumor progression: clonal evolution of human stem cell populations.
Calabrese P, Tavaré S, Shibata D. Calabrese P, et al. Am J Pathol. 2004 Apr;164(4):1337-46. doi: 10.1016/S0002-9440(10)63220-8. Am J Pathol. 2004. PMID: 15039221 Free PMC article. - High incidence of epithelial cancers in mice deficient for DNA polymerase delta proofreading.
Goldsby RE, Hays LE, Chen X, Olmsted EA, Slayton WB, Spangrude GJ, Preston BD. Goldsby RE, et al. Proc Natl Acad Sci U S A. 2002 Nov 26;99(24):15560-5. doi: 10.1073/pnas.232340999. Epub 2002 Nov 12. Proc Natl Acad Sci U S A. 2002. PMID: 12429860 Free PMC article. - The Genetic Basis of Mutation Rate Variation in Yeast.
Gou L, Bloom JS, Kruglyak L. Gou L, et al. Genetics. 2019 Feb;211(2):731-740. doi: 10.1534/genetics.118.301609. Epub 2018 Nov 30. Genetics. 2019. PMID: 30504363 Free PMC article. - Mathematical modeling for mutator phenotype and clonal selection advantage in the risk analysis of lung cancer.
Li L, Zhao T, He X, Yang X, Tian T, Zhang X. Li L, et al. Theory Biosci. 2022 Sep;141(3):261-272. doi: 10.1007/s12064-022-00371-z. Epub 2022 Jun 4. Theory Biosci. 2022. PMID: 35665446 - What does physics have to do with cancer?
Michor F, Liphardt J, Ferrari M, Widom J. Michor F, et al. Nat Rev Cancer. 2011 Aug 18;11(9):657-70. doi: 10.1038/nrc3092. Nat Rev Cancer. 2011. PMID: 21850037 Free PMC article. Review.
References
- Fisher J C. Nature (London) 1958;181:651–652. - PubMed
- Cairns J. Nature (London) 1975;255:197–200. - PubMed
- Knudson A G. J Cancer Res Clin Oncol. 1996;122:135–140. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources