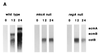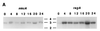Developmental signal transduction pathways uncovered by genetic suppressors - PubMed (original) (raw)
Comparative Study
Developmental signal transduction pathways uncovered by genetic suppressors
G Shaulsky et al. Proc Natl Acad Sci U S A. 1996.
Abstract
We have found conditions for saturation mutagenesis by restriction enzyme mediated integration that result in plasmid tagging of disrupted genes. Using this method we selected for mutations in genes that act at checkpoints downstream of the intercellular signalling system that controls encapsulation in Dictyostelium discoideum. One of these genes, mkcA, is a member of the mitogen-activating protein kinase cascade family while the other, regA is a novel bipartite gene homologous to response regulators in one part and to cyclic nucleotide phosphodiesterases in the other part. Disruption of either of these genes results in partial suppression of the block to spore formation resulting from the loss of the prestalk genes, tagB and tagC. The products of the tag genes have conserved domains of serine protease attached to ATP-driven transporters, suggesting that they process and export peptide signals. Together, these genes outline an intercellular communication system that coordinates organismal shape with cellular differentiation during development.
Figures
Figure 1
Sequence analysis of mkcA and_regA_. (A) Sequenced regions of_mkcA_ and regA genomic DNA are shown. Coding regions are indicated in boxes, and V shapes indicate introns. Plasmid (4.5 kb, not to scale) insertion sites (IS) in the respective mutant strains are indicated as triangles. Solid box in_mkcA_ is the kinase homology domain. Thatched boxes in_regA_ encode the response regulator homology domain, and checked boxes encode the cyclic nucleotide phosphodiesterase homology domain. Gray boxes under the genes indicate cDNA probes. (B) Sequence similarity between the putative protein kinase domain of_mkcA_ and those of the mitogen-activating protein (MAP) kinase cascade genes PAK65 (Swiss-Prot Protein Sequence Data Bank no. P35465P35465) and STE20 (GenBank accession no. L04655L04655). (C) Sequence similarity between the putative response regulator domain of regA and those of the bacterial response regulators cheY [National Center for Biotechnology Information (NCBI) no. 145525], ntrC (Swiss-Prot Protein Sequence Data Bank no. P10576P10576), and_pleD_ (NCBI no. 1119215). (D) Sequence similarity between the putative cyclic nucleotide phosphodiesterase domain of regA and those of a rat phosphodiesterase (r.PDE; NCBI no. 436012) and a bovine calmodulin stimulated phosphodiesterase (b.PDE; NCBI no. 533781). Conserved regions are underlined, and amino acid numbers are indicated on the left.
Figure 2
Expression of cell type-specific genes in_mkcA_ null or regA null strains. (A) RNA from wild-type cells and from mutants was prepared at 0, 12, and 24 h of development as indicated. Northern blots were hybridized with cotB, a prespore-specific gene probe and with ecmA and ecmB, two prestalk-specific gene probes. (B) Wild-type or mutant cells were developed to early culmination and subjected to in situ RNA hybridization with riboprobes for ecmA,ecmB, and cotB as indicated. Arrow shows the lower cup. (Bar = 0.2 mm.)
Figure 2
Expression of cell type-specific genes in_mkcA_ null or regA null strains. (A) RNA from wild-type cells and from mutants was prepared at 0, 12, and 24 h of development as indicated. Northern blots were hybridized with cotB, a prespore-specific gene probe and with ecmA and ecmB, two prestalk-specific gene probes. (B) Wild-type or mutant cells were developed to early culmination and subjected to in situ RNA hybridization with riboprobes for ecmA,ecmB, and cotB as indicated. Arrow shows the lower cup. (Bar = 0.2 mm.)
Figure 3
Regulation and cell type specificity of_mkcA_ and regA expression. (A) RNA was prepared from developing wild-type cells at 4-h intervals and analyzed by Northern blot analysis using cDNA riboprobes for_mkcA_ or regA (Fig. 1) as indicated. Time (h) is indicated above the lanes; size (kb) is indicated between blots. (B) Wild-type cells were developed to the finger stage or to early culmination and subjected to in situ RNA hybridization with cDNA riboprobes for mkcA or_regA_ as indicated. (Bar = 0.2 mm.)
Figure 3
Regulation and cell type specificity of_mkcA_ and regA expression. (A) RNA was prepared from developing wild-type cells at 4-h intervals and analyzed by Northern blot analysis using cDNA riboprobes for_mkcA_ or regA (Fig. 1) as indicated. Time (h) is indicated above the lanes; size (kb) is indicated between blots. (B) Wild-type cells were developed to the finger stage or to early culmination and subjected to in situ RNA hybridization with cDNA riboprobes for mkcA or_regA_ as indicated. (Bar = 0.2 mm.)
Similar articles
- Signal transduction pathways leading to spore differentiation in Dictyostelium discoideum.
Anjard C, Zeng C, Loomis WF, Nellen W. Anjard C, et al. Dev Biol. 1998 Jan 15;193(2):146-55. doi: 10.1006/dbio.1997.8804. Dev Biol. 1998. PMID: 9473320 - A multidrug resistance transporter/serine protease gene is required for prestalk specialization in Dictyostelium.
Shaulsky G, Kuspa A, Loomis WF. Shaulsky G, et al. Genes Dev. 1995 May 1;9(9):1111-22. doi: 10.1101/gad.9.9.1111. Genes Dev. 1995. PMID: 7744252 - Precocious sporulation and developmental lethality in yelA null mutants of Dictyostelium.
Osherov N, Wang N, Loomis WF. Osherov N, et al. Dev Genet. 1997;20(4):307-19. doi: 10.1002/(SICI)1520-6408(1997)20:4<307::AID-DVG2>3.0.CO;2-B. Dev Genet. 1997. PMID: 9254905 - Intercellular signaling. A kinase for cell-fate determination?
Briscoe C, Firtel RA. Briscoe C, et al. Curr Biol. 1995 Mar 1;5(3):228-31. doi: 10.1016/s0960-9822(95)00045-5. Curr Biol. 1995. PMID: 7780726 Review. - A model for cGMP signal transduction in Dictyostelium in perspective of 25 years of cGMP research.
Bosgraaf L, Van Haastert PJ. Bosgraaf L, et al. J Muscle Res Cell Motil. 2002;23(7-8):781-91. doi: 10.1023/a:1024431813040. J Muscle Res Cell Motil. 2002. PMID: 12952076 Review.
Cited by
- Evolutionary crossroads in developmental biology: Dictyostelium discoideum.
Schaap P. Schaap P. Development. 2011 Feb;138(3):387-96. doi: 10.1242/dev.048934. Development. 2011. PMID: 21205784 Free PMC article. Review. - Initial cell type choice in Dictyostelium.
Jang W, Gomer RH. Jang W, et al. Eukaryot Cell. 2011 Feb;10(2):150-5. doi: 10.1128/EC.00219-10. Epub 2010 Dec 10. Eukaryot Cell. 2011. PMID: 21148754 Free PMC article. Review. - Identification of a novel type of cGMP phosphodiesterase that is defective in the chemotactic stmF mutants.
Meima ME, Biondi RM, Schaap P. Meima ME, et al. Mol Biol Cell. 2002 Nov;13(11):3870-7. doi: 10.1091/mbc.e02-05-0285. Mol Biol Cell. 2002. PMID: 12429831 Free PMC article. - TgrC1 Has Distinct Functions in Dictyostelium Development and Allorecognition.
Wang Y, Shaulsky G. Wang Y, et al. PLoS One. 2015 Apr 20;10(4):e0124270. doi: 10.1371/journal.pone.0124270. eCollection 2015. PLoS One. 2015. PMID: 25894230 Free PMC article. - Transcriptional transitions during Dictyostelium spore germination.
Xu Q, Ibarra M, Mahadeo D, Shaw C, Huang E, Kuspa A, Cotter D, Shaulsky G. Xu Q, et al. Eukaryot Cell. 2004 Oct;3(5):1101-10. doi: 10.1128/EC.3.5.1101-1110.2004. Eukaryot Cell. 2004. PMID: 15470238 Free PMC article.
References
- Shaulsky G, Kuspa A, Loomis W F. Genes Dev. 1995;9:1111–1122. - PubMed
- Adames N, Blundell K, Ashby M N, Boone C. Science. 1995;270:464–467. - PubMed
- McGarth J P, Varshavsky A. Nature (London) 1989;340:400–404. - PubMed
- Michaelis S. Semin Cell Biol. 1993;4:17–27. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases