Visualization of the peroxisomal compartment in living mammalian cells: dynamic behavior and association with microtubules - PubMed (original) (raw)
Visualization of the peroxisomal compartment in living mammalian cells: dynamic behavior and association with microtubules
E A Wiemer et al. J Cell Biol. 1997.
Abstract
Peroxisomes in living CV1 cells were visualized by targeting the green fluorescent protein (GFP) to this subcellular compartment through the addition of a COOH-terminal peroxisomal targeting signal 1 (GFP-PTS1). The organelle dynamics were examined and analyzed using time-lapse confocal laser scanning microscopy. Two types of movement could be distinguished: a relatively slow, random, vibration-like movement displayed by the majority (approximately 95%) of the peroxisomes, and a saltatory, fast directional movement displayed by a small subset (approximately 5%) of the peroxisomes. In the latter instance, peak velocities up to 0.75 micron/s and sustained directional velocities up to 0.45 micron/s over 11.5 microns were recorded. Only the directional type of motion appeared to be energy dependent, whereas the vibrational movement continued even after the cells were depleted of energy. Treatment of cells, transiently expressing GFP-PTS1, with microtubule-destabilizing agents such as nocodazole, vinblastine, and demecolcine clearly altered peroxisome morphology and subcellular distribution and blocked the directional movement. In contrast, the microtubule-stabilizing compound paclitaxel, or the microfilament-destabilizing drugs cytochalasin B or D, did not exert these effects. High resolution confocal analysis of cells expressing GFP-PTS1 and stained with anti-tubulin antibodies revealed that many peroxisomes were associated with microtubules. The GFP-PTS1-labeled peroxisomes were found to distribute themselves in a stochastic, rather than ordered, manner to daughter cells at the time of mitosis.
Figures
Figure 1
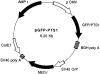
GFP–PTS1 expression construct. A mutant version (S65T) of the GFP cDNA was cloned into the vector pcDNA3 as described in Materials and Methods. The resulting plasmids either expressed GFP (pGFP) alone or GFP onto which a peroxisomal type I targeting signal (Ser-Lys-Leu-COOH) was fused at its COOH-terminal end (pGFP-PTS1). Depicted is pGFP-PTS1 containing the GFP–PTS1 construct under control of the human cytomegalovirus intermediate early promoter/enhancer region (pCMV). BGH poly A, bovine growth hormone polyadenylation signal; SV40 O/P, SV40 origin and early promoter; NEO r, neomycin-resistance gene; SV40 poly A, SV40 T-antigen polyadenylation signal; Col E1, col E1 origin of replication; AMP r, β-lactamase gene.
Figure 2
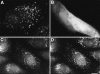
Subcellular localization of GFP and GFP–PTS1 in CV1 cells. CV1 cells were cultured on coverslips, transfected with pGFP and pGFP-PTS1. After 48 h the cells were examined directly for GFP–PTS1 (A) or GFP (B) expression. Alternatively, pGFP-PTS1– transfected CV1 cells were fixed, permeabilized, and processed for immunofluorescence. In a double-labeling experiment, the cells were incubated with rabbit anti–PMP70, and subsequently with goat anti–rabbit IgG conjugated to rhodamine. The cells were monitored for the subcellular localization of GFP–PTS1 (C) in the FITC channel and PMP70 (D) in the rhodamine channel. Bar, 10 μm.
Figure 3
Time-lapse motion analysis of the dynamics of peroxisomes in living CV1 cells expressing GFP–PTS1. A representative single cell was selected, and single-frame confocal images were recorded at 6.4-s intervals for a total of 500 frames. 20 consecutive frames were chosen for motion analysis. (A) Three confocal images from the series are shown at time = 0 (frame 001), time = 64 s (frame 010), and time = 128 s (frame 020). Most peroxisomes displayed a slow, random movement within a limited space, leaving their overall distribution relatively unchanged after ∼2 min. Bar, 10 μm. (B) Plot of the peak velocities of 85 individual peroxisomes (∼50% of the cell population) observed over 20 consecutive frames revealed two distinct types of motion. The majority (95%) exhibited a localized, random type of Brownian motion with peak velocities of <0.2 μm/s. A smaller number (∼5%) exhibited a faster motion characterized by peak velocities of >0.2 μm/s and as great as 0.68 μm/s. (C) Plot of the mean average velocity of 85 individual peroxisomes over 20 consecutive frames also showed two distinct types of motion. The majority exhibited a localized, random type of Brownian motion with an average velocity of <0.05 μm/s, while the faster moving peroxisomes displayed an average velocity of >0.05 μm/s. (D) Vector diagram plot tracking the motion of eight representative individual peroxisomes (A–H) corresponding to those marked in C. A–E exhibited very little net displacement and were characteristic of the majority of peroxisomes. Peroxisome F exhibited sporadic motion, G was relatively stationary for several frames before exhibiting rapid directional motion, and H had a more continuous, rapid directional motion over the time course (1 pixel = 0.13 μm).
Figure 4
Time-lapse analysis of the effects of paclitaxel and nocodazole on the dynamics of peroxisomes in living CV1 cells expressing GFP–PTS1. Representative single cells were selected and confocal images were taken at 6.4-s intervals for a total of 500 frames. 20 frames for each condition were chosen for motion analysis. (A) Three confocal images of a cell treated with paclitaxel at time = 0 (frame 001), time = 64 s (frame 010), and time = 128 s (frame 020) are shown. The overall distribution of peroxisomes was similar to that in normal cells. (B) Plot of the peak velocity of 85 individual peroxisomes (∼50% of the cell population) over 20 consecutive frames appeared similar to that found in normal cells (Fig. 3_B_). (C) Plot of the mean average velocity of 85 individual peroxisomes over 20 consecutive frames appeared similar to that found in normal cells (Fig. 3_C_). (D) Vector diagram plot tracking the motion of eight representative peroxisomes (A–H) corresponding to those marked in C. A–D were relatively motionless, while E and F exhibited sporadic motion and G and H exhibited more continuous rapid motion (1 pixel = 0.13 μm). The average velocities and mean changes in position were similar to those found in normal cells (Fig. 3_D_). (E) Three frames from a series as in A of a cell treated with nocodazole showed an abnormal clustering of peroxisomes. (F) Plot of the peak velocity of 85 individual peroxisomes (∼50% of the cell population) over 20 consecutive frames appeared abnormal. All peroxisomes exhibited only a localized, random type of Brownian motion with none exceeding a peak velocity 0.2 μm/s. (G) Plot of the mean average velocity showed that only a few peroxisomes exceeded 0.05 μm/s. (H) Vector diagram plot tracking the motion of eight representative peroxisomes (A–H) corresponding to those marked in G. None of the peroxisomes exhibited rapid motion or displacements >0.5 μm over the time course (1 pixel = 0.13 μm).
Figure 5
High resolution confocal analysis of the movement of individual peroxisomes. (A) Five frames (000–004) taken at 6.4-s intervals showed the rapid unidirectional movement of a single peroxisome (arrows). The peak velocity of this peroxisome was 0.75 μm/s with an average velocity measured over 11.5 μm of 0.45 μm/s. (B) 15 frames (000–014) taken at 6.4-s intervals showed the bidirectional movement of a single elongated peroxisome. In frames 001–006, a peroxisome could be seen making a rapid looping motion and coming to a complete stop (arrows). In frames 006 and 007, the long axis of the peroxisome was oriented in the vertical dimension. In frames 007–014, the same peroxisome reversed course and returned along the same path with a similar velocity (arrows). Bars, 10 μm.
Figure 6
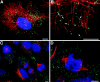
Relationship of peroxisomes to microtubules and peroxisomal distribution during mitosis. Triple fluorescence confocal imaging of CV1 cells expressing GFP in peroxisomes (green), propidium iodide staining of the nucleus (pseudo-colored blue), and immunolabeling of microtubules using mouse anti–β-tubulin followed by goat anti–mouse IgG-CY5 (pseudo-colored red). (A) Low power image of the overall distribution of peroxisomes related to microtubules. (B) Higher power image of the same cell showing the close association of many of the individual peroxisomes (arrows) with individual microtubules. (C) Low power image of cells at metaphase and anaphase stages of mitosis. (D) Higher power image of an individual cell during anaphase stage of mitosis showing the random distribution of peroxisomes. Bars, 10 μm.
Similar articles
- Interaction of microtubules with peroxisomes. Tubular and spherical peroxisomes in HepG2 cells and their alterations induced by microtubule-active drugs.
Schrader M, Burkhardt JK, Baumgart E, Lüers G, Spring H, Völkl A, Fahimi HD. Schrader M, et al. Eur J Cell Biol. 1996 Jan;69(1):24-35. Eur J Cell Biol. 1996. PMID: 8825021 - Visualization of peroxisomes in living plant cells reveals acto-myosin-dependent cytoplasmic streaming and peroxisome budding.
Jedd G, Chua NH. Jedd G, et al. Plant Cell Physiol. 2002 Apr;43(4):384-92. doi: 10.1093/pcp/pcf045. Plant Cell Physiol. 2002. PMID: 11978866 - Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: dynamic morphology and actin-dependent movement.
Mano S, Nakamori C, Hayashi M, Kato A, Kondo M, Nishimura M. Mano S, et al. Plant Cell Physiol. 2002 Mar;43(3):331-41. doi: 10.1093/pcp/pcf037. Plant Cell Physiol. 2002. PMID: 11917088 - Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants.
Mathur J, Mathur N, Hülskamp M. Mathur J, et al. Plant Physiol. 2002 Mar;128(3):1031-45. doi: 10.1104/pp.011018. Plant Physiol. 2002. PMID: 11891258 Free PMC article. - Targeted fluorescent probes in peroxisome function.
Dansen TB, Pap EHW, Wanders RJ, Wirtz KW. Dansen TB, et al. Histochem J. 2001 Feb;33(2):65-9. doi: 10.1023/a:1017927728892. Histochem J. 2001. PMID: 11432641 Review.
Cited by
- A Microfluidic Device for Modulation of Organellar Heterogeneity in Live Single Cells.
Wada KI, Hosokawa K, Ito Y, Maeda M. Wada KI, et al. Anal Sci. 2021 Mar 10;37(3):499-503. doi: 10.2116/analsci.20SCP11. Epub 2020 Dec 4. Anal Sci. 2021. PMID: 33281140 - Quantitative analysis of peroxisome tracks using a Hidden Markov Model.
Svensson CM, Reglinski K, Schliebs W, Erdmann R, Eggeling C, Figge MT. Svensson CM, et al. Sci Rep. 2023 Nov 11;13(1):19694. doi: 10.1038/s41598-023-46812-7. Sci Rep. 2023. PMID: 37951993 Free PMC article. - Single-motor and multi-motor motility properties of kinesin-6 family members.
Poulos A, Budaitis BG, Verhey KJ. Poulos A, et al. Biol Open. 2022 Oct 15;11(10):bio059533. doi: 10.1242/bio.059533. Epub 2022 Oct 14. Biol Open. 2022. PMID: 36178151 Free PMC article. - Miro GTPase domains regulate the assembly of the mitochondrial motor-adaptor complex.
Davis K, Basu H, Izquierdo-Villalba I, Shurberg E, Schwarz TL. Davis K, et al. Life Sci Alliance. 2022 Oct 27;6(1):e202201406. doi: 10.26508/lsa.202201406. Print 2023 Jan. Life Sci Alliance. 2022. PMID: 36302649 Free PMC article. - Peroxisomes in dental tissues of the mouse.
Stelzig I, Karnati S, Valerius KP, Baumgart-Vogt E. Stelzig I, et al. Histochem Cell Biol. 2013 Oct;140(4):443-62. doi: 10.1007/s00418-013-1131-8. Epub 2013 Aug 28. Histochem Cell Biol. 2013. PMID: 23982811
References
- Allan V. Organelle movement. Dynactin: portrait of a dynein regulator. Curr Biol. 1994;4:1000–1002. - PubMed
- Baumann O, Murphy DB. Microtubule-associated movement of mitochondria and small particles in Acanthamoeba castellanii. . Cell Motil Cytoskeleton. 1995;32:305–317. - PubMed
- Birky CW. The partitioning of cytoplasmic organelles at cell division. Int Rev Cytol. 1983;15:49–89. - PubMed
- Cole NB, Lippincott-Schwartz J. Organization of organelles and membrane traffic by microtubules. Curr Opin Cell Biol. 1995;7:55–64. - PubMed
- Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R56 DK041737/DK/NIDDK NIH HHS/United States
- RR04050/RR/NCRR NIH HHS/United States
- R37 DK041737/DK/NIDDK NIH HHS/United States
- NS14718/NS/NINDS NIH HHS/United States
- DK41737/DK/NIDDK NIH HHS/United States
- P41 RR004050/RR/NCRR NIH HHS/United States
- R01 NS014718/NS/NINDS NIH HHS/United States
- R01 DK041737/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources