Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry - PubMed (original) (raw)
Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry
G Neubauer et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Here we report the rapid identification of the proteins of the spliceosomal U1 small nuclear ribonucleoprotein (snRNP) from the yeast Saccharomyces cerevisiae by searching mass spectrometric data in genomic sequence databases. The U1 snRNP, containing a histidine-tagged 70K protein, was isolated from cell extracts by anti m3G-cap immunoaffinity and subsequent nickel nitrilotriacetic acid chromatography. A U1 snRNP fraction containing 20 proteins was obtained. Further purification by glycerol gradient centrifugation identified nine U1 snRNP specific and six common proteins. The U1 snRNP proteins were partially sequenced by nanoelectrospray mass spectrometry, and their genes were identified in the data base via multiple peptide sequence tags. Apart from the already known common proteins D1, D3, F, and G, the D2 and E homologs were also identified. The same six common proteins were detected in core U2 snRNP, which was purified and analyzed separately. The biochemical association of these six proteins with yeast snRNPs is shown here for the first time. Intriguingly, the Sm B/B' homolog was not detected. In addition to the well characterized yeast U1 specific proteins [U1-70K (Snp1p), U1-A (Mud1p), Prp39p, and Prp40p] the homolog of the U1-C protein was identified together with four additional novel U1 specific proteins, which are not found in mammalian U1. This is the first time that the components of a multiprotein complex from an organism with a sequenced genome have been characterized by mass spectrometry. The technique should be applicable to any protein complex that can be biochemically purified from an organism whose genome is known.
Figures
Figure 1
Purification of U1 snRNPs from S. cerevisiae. (A) Silver staining of snRNAs eluted from anti-m3G-cap (m7G eluate) and Ni-NTA affinity columns (Ni-NTA eluate). The positions of U1 and U5 (long and short form) RNAs are indicated. (B) Proteins of the total snRNPs (m7G eluate) were stained with silver. The proteins of the Ni-NTA eluate were stained with Coomassie blue and then partially sequenced by mass spectrometry. We estimate the protein amounts to be between 5 and less than 1 picomole. Proteins of 216, 115, and 42 kDa are associated with U5 snRNPs (see gradient centrifugation, C and D). The two proteins of 36 and 38 kDa (asterisks) are contaminants. All of the other proteins are U1 snRNP-associated. The band of 69 kDa contained two U1 specific proteins, Prp39p and Prp40p. The U1-C protein was identified in two bands of 32 and 31 kDa. Six yeast common snRNP proteins were identified in the low molecular weight region between 9 and 18 kDa. Molecular weights are indicated in kilodaltons. (C) U1 and U5 snRNPs were separated on a glycerol gradient, and the snRNAs of the corresponding fractions were stained with silver. U1 and U5 snRNAs are indicated. 18S U1 snRNP peaks in fractions 16 and 17, whereas 15S U5 snRNP peaks in fractions 13 and 14. The RNA in the U3 region (asterisk), which co-migrates with U5, was not present in the preparation used for the sequencing of proteins. (D) Proteins of separated U1 and U5 snRNPs were stained with silver. Proteins of 216, 115, and 42 kDa co-migrate with U5 RNA. All of the other proteins co-migrate with U1 RNA. The common proteins of molecular masses ranging from 9 to 18 kDa co-migrate with U1 as well as U5 RNAs. U1 snRNP proteins are indicated on the right, U5 proteins on the left. The band at 49 kDa (asterisk) is a RNase inhibitor.
Figure 2
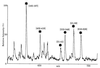
Identification of the 15 kDa band of Fig. 1_B_ by mass spectrometry. (A) Part of the mass spectrum of the peptide mixture obtained after digestion of the protein band. (B) Fragmentation of the peak at m/z 756.9 in part A. Mass spectrum of the fragments obtained by scanning the third quadrupole Q3. The prominent fragments in the high molecular weight region are spaced by amino acid molecular weights and reveal the partial sequence LEEL as shown by the arrows (note that the isobaric amino acids leucine and isoleucine are indistinguishable by our method). The sequence stretch, together with its starting mass, its end mass, and the molecular weight of the peptide are entered into a database searching program (
peptidesearch
) where they are converted to a peptide sequence tag. Search of the tag in a nonredundant database uniquely retrieves the sequence AELEELEEFEFK from gene L9328.5 (GenBank). The predicted C-terminal or Y" ion fragments (4, 22) of this peptide are marked in the spectrum and verify the complete sequence. (C) Protein sequence of gene L9328.5. Five peptides were partially sequenced that identified peptides from the gene as indicated by underlining (those within the mass range are also marked by bullets in A). The partial sequence deduced from B is in gray and the complete peptide is boxed.
Figure 3
Identification of Prp39p and Prp40p in the 69 kDa band of Fig. 1_B_. Part of the tandem mass spectrum of peptides extracted from the band. The marked peaks have been sequenced, and their peptide sequence tags searched in the data base. They mapped to the indicated sequence range in Prp39p (stars) or Prp40p (bullets). The sequenced peptides cover 106 and 41 amino acids for Prp39p and Prp40p, respectively; thus identification of both proteins is certain.
Figure 4
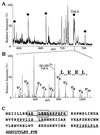
(A) Alignment of the yeast and human proteins Sm E. The Sm motifs 1 (boxed in light gray) and Sm motif 2 (boxed in dark gray) are indicated. (B) Alignment of the yeast and human proteins Sm D2. (C) Alignment of yeast and human U1-C proteins. The four metal binding residues of a putative zinc finger motif are boxed in gray. Printed in boldface letters are the peptides which were sequenced by mass spectrometry. SC, S. cerevisiae; HS, Homo sapiens.
Similar articles
- A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins.
Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot HV, Mann M, Séraphin B, Rosbash M, Lührmann R, Fabrizio P. Gottschalk A, et al. RNA. 1998 Apr;4(4):374-93. RNA. 1998. PMID: 9630245 Free PMC article. - The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins.
Nelissen RL, Will CL, van Venrooij WJ, Lührmann R. Nelissen RL, et al. EMBO J. 1994 Sep 1;13(17):4113-25. doi: 10.1002/j.1460-2075.1994.tb06729.x. EMBO J. 1994. PMID: 8076607 Free PMC article. - Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p.
Lockhart SR, Rymond BC. Lockhart SR, et al. Mol Cell Biol. 1994 Jun;14(6):3623-33. doi: 10.1128/mcb.14.6.3623-3633.1994. Mol Cell Biol. 1994. PMID: 8196608 Free PMC article. - Identification and characterization of a yeast homolog of U1 snRNP-specific protein C.
Tang J, Abovich N, Fleming ML, Seraphin B, Rosbash M. Tang J, et al. EMBO J. 1997 Jul 1;16(13):4082-91. doi: 10.1093/emboj/16.13.4082. EMBO J. 1997. PMID: 9233817 Free PMC article.
Cited by
- Arrested yeast splicing complexes indicate stepwise snRNP recruitment during in vivo spliceosome assembly.
Tardiff DF, Rosbash M. Tardiff DF, et al. RNA. 2006 Jun;12(6):968-79. doi: 10.1261/rna.50506. Epub 2006 Apr 17. RNA. 2006. PMID: 16618970 Free PMC article. - Uncovering the rules for protein-protein interactions from yeast genomic data.
Wang J, Li C, Wang E, Wang X. Wang J, et al. Proc Natl Acad Sci U S A. 2009 Mar 10;106(10):3752-7. doi: 10.1073/pnas.0806427106. Epub 2009 Feb 23. Proc Natl Acad Sci U S A. 2009. PMID: 19237561 Free PMC article. - Inventories to insights.
Aitchison JD, Galitski T. Aitchison JD, et al. J Cell Biol. 2003 May 12;161(3):465-9. doi: 10.1083/jcb.200302041. J Cell Biol. 2003. PMID: 12743099 Free PMC article. Review. - Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex.
Panigrahi AK, Gygi SP, Ernst NL, Igo RP Jr, Palazzo SS, Schnaufer A, Weston DS, Carmean N, Salavati R, Aebersold R, Stuart KD. Panigrahi AK, et al. Mol Cell Biol. 2001 Jan;21(2):380-9. doi: 10.1128/MCB.21.2.380-389.2001. Mol Cell Biol. 2001. PMID: 11134327 Free PMC article. - Genetic and biochemical analysis of yeast and human cap trimethylguanosine synthase: functional overlap of 2,2,7-trimethylguanosine caps, small nuclear ribonucleoprotein components, pre-mRNA splicing factors, and RNA decay pathways.
Hausmann S, Zheng S, Costanzo M, Brost RL, Garcin D, Boone C, Shuman S, Schwer B. Hausmann S, et al. J Biol Chem. 2008 Nov 14;283(46):31706-18. doi: 10.1074/jbc.M806127200. Epub 2008 Sep 5. J Biol Chem. 2008. PMID: 18775984 Free PMC article.
References
- Patterson S D, Aebersold R. Electrophoresis. 1995;16:791–814. - PubMed
- Fenn J B, Mann M, Meng C K, Wong S F, Whitehouse C M. Science. 1989;246:64–71. - PubMed
- Mann M, Wilm M. Trends Biochem Sci. 1995;20:219–223. - PubMed
- Mann M, Wilm M. Anal Chem. 1994;66:4390–4399. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous