Characterization of the chicken beta-globin insulator - PubMed (original) (raw)
Characterization of the chicken beta-globin insulator
J H Chung et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Insulators, first identified in Drosophila, are DNA sequence elements that shield a promoter from nearby regulatory elements. We have previously reported that a DNA sequence at the 5' end of the chicken beta-globin locus can function as an insulator. It is capable of shielding a reporter gene from the activating effects of a nearby mouse beta-globin locus control region element in the human erythroleukemic cell line K562. In this report, we show that most of the insulating activity lies in a 250-bp CpG island (core element), which contains the constitutive DNase I-hypersensitive site (5'HS4). DNA binding assays with the core sequence reveal a complex protein binding pattern. The insulating activity of the core element is multiplied when tandem copies are used. Although CpG islands are often associated with promoters of housekeeping genes, we find little evidence that the core element is a promoter. Furthermore, the insulator differs from a promoter in its ability to block the locus control region effect directionally.
Figures
Figure 5
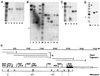
Analysis of DNA methylation in the insulator region. In each of the panels (A_–_D), the sensitivity of the parent fragments illustrated in E to digestion by methylation-sensitive restriction enzymes is shown. In A, a 454-bp parent fragment was mock-digested (lane 1), digested with the methylation-sensitive enzymes _Hpa_II (lane 2), _Sma_I (lane 4), _Hha_I (lane 5), and _Aci_I (lane 6), or digested with the methylation-insensitive isoschizomer of _Hpa_II–_Msp_I (lane 3). In lanes 1–4 of B, 1469- and 1219-bp parent fragments were mock-digested (lane 1), digested with the methylation-sensitive enzymes _Hpa_II (lane 2) and _Ava_I (lane 4), or digested with the methylation-insensitive isoschizomer of _Hpa_II–Msp I (lane 3). In lanes 5–9 of B, an 893-bp parent fragment was mock-digested (lane 5), digested with the methylation-sensitive enzymes _Hpa_II (lane 6), _Fsp_I (lane 8), and _Hha_I (lane 9), or digested with the methylation-insensitive isoschizomer of _Hpa_II–_Msp_I (lane 3). In C, a 503-bp parent fragment was mock-digested (lane 1), digested with the methylation-sensitive enzymes _Hha_I (lane 2) and _Hpa_II (lane 4), or digested with the methylation-insensitive isoschizomer of _Hpa_II–_Msp_I (lane 3). In D, a 1017-bp parent fragment was mock-digested (lane 1) or digested with the methylation-sensitive enzymes _Sma_I (lane 2) and _Hae_II (lane 3). In A_–_D, the arrowhead indicates the position of the 500-bp fragment of the 100-bp ladder in lane M. The results of these experiments are summarized qualitatively in E. +, Full methylation; +/−, partial methylation; and −, absence of detectable methylation. The boxed region delineates the position of the 1.2-kb insulator; stippling indicates the position of the core.
Figure 1
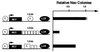
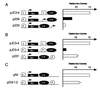
Localization of the insulator activity. Three DNA fragments containing various regions of the intact 1.2-kb fragment (I, intact fragment; C1, 1 kb of the 3′ region; C2, 600 bp of the 3′ region; and C3, 650 bp of the 5′ region) were inserted on both sides of the Aγ-neo reporter gene. These constructs were stably transfected into K562 cells and G418-resistant colonies were selected and counted 2 to 3 weeks later as described previously (5). The number of G418-resistant colonies for pJC3-4 was set to 1.0.
Figure 2
Testing the insulating activity of the core element. Six copies of the 250-bp fragment containing 5′HS4 (p179) or two copies of the fragment plus a 950-bp fragment from λ phage DNA (p176) were inserted between the LCR and the Aγ-neo reporter gene. The number of G418-resistant colonies was determined as in Fig. 1, and the number obtained for pJC5-4, which contains one copy of the intact insulator on both sides of the reporter gene, was set to 1.0.
Figure 7
Comparing the position dependence of insulator activity with that of the Aγ-globin promoter. A 559-bp fragment containing the Aγ-globin promoter (A) or the full-length insulator fragment (B) was inserted either between the LCR and the reporter (p239 and pJC5-4, respectively) or on the outside of the LCR (p233 and p231, respectively). In C, a construct that contains the insulator inserted on the outside of the LCR is compared with an otherwise identical construct in which no insulator has been inserted. For each construct, the number of G418-resistant colonies was determined as in Fig. 1 and normalized to pJC3-4 (A and B) or pNI (C) as indicated. The data presented are the average of two to four independent experiments.
Figure 3
The sequence of the insulator core element. The heavy bars indicate the footprinted regions observed for both the coding (upper) and noncoding (lower) strand. The footprinted regions are numbered (I–IV) according to their positions on each strand; note that the positions of the fourth footprinted regions on the upper and lower strands do not coincide. The 5′ boundaries of the overlapping oligonucleotides used as competitors in DNase I footprint analysis (Fig. 4) are indicated as vertical lines. Oligonucleotide 42/43 is 25 bp long; all of the remaining oligonucleotides are 40 bp long. The extended footprint II and a hypersensitive site seen in the presence of the competing oligonucleotide 34/35 are shown as striped bars and a vertical arrow, respectively. The vertical arrow also indicates the boundary between the footprints IIA and IIB. The potential binding sites for Sp1, the yeast α2 repressor, and suppressor of hairy wing are underlined.
Figure 4
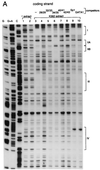
DNase I footprint analysis of the core element. DNA binding reactions were incubated for 20 min at 25°C with K562 nuclear extract. (A) Coding strand analysis. The four most prominent footprints are labeled I–IV. The oligonucleotide competitor, where used, is indicated above the lanes. See Fig. 3 for the sequence compositions of the competitors as well as a summary of the footprint analysis. To control for specificity, oligonucleotide competitors with GATA1 and Sp1 binding sites were also used (lanes 9 and 10, respectively). (B) Noncoding strand analysis. In addition to the four observed footprints, the location of the canonical Sp1 binding site is also indicated.
Figure 4
DNase I footprint analysis of the core element. DNA binding reactions were incubated for 20 min at 25°C with K562 nuclear extract. (A) Coding strand analysis. The four most prominent footprints are labeled I–IV. The oligonucleotide competitor, where used, is indicated above the lanes. See Fig. 3 for the sequence compositions of the competitors as well as a summary of the footprint analysis. To control for specificity, oligonucleotide competitors with GATA1 and Sp1 binding sites were also used (lanes 9 and 10, respectively). (B) Noncoding strand analysis. In addition to the four observed footprints, the location of the canonical Sp1 binding site is also indicated.
Figure 6
Testing the core element for promoter activity. Three reporter genes, including pGL2-basic, which has no promoter, pGL2-promoter, which has the SV40 early promoter, and p265, which has the core element inserted in place of the SV40 promoter, were transiently transfected into K562 cells, and luciferase activity was measured 48 hr later.
Similar articles
- The 5'HS4 core element of the human beta-globin locus control region is required for high-level globin gene expression in definitive but not in primitive erythropoiesis.
Navas PA, Peterson KR, Li Q, McArthur M, Stamatoyannopoulos G. Navas PA, et al. J Mol Biol. 2001 Sep 7;312(1):17-26. doi: 10.1006/jmbi.2001.4939. J Mol Biol. 2001. PMID: 11545582 - The mouse beta-globin locus control region: hypersensitive sites 3 and 4.
Jiménez G, Gale KB, Enver T. Jiménez G, et al. Nucleic Acids Res. 1992 Nov 11;20(21):5797-803. doi: 10.1093/nar/20.21.5797. Nucleic Acids Res. 1992. PMID: 1454540 Free PMC article. - Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities.
Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, Gaszner M, Felsenfeld G. Recillas-Targa F, et al. Proc Natl Acad Sci U S A. 2002 May 14;99(10):6883-8. doi: 10.1073/pnas.102179399. Proc Natl Acad Sci U S A. 2002. PMID: 12011446 Free PMC article. - Use of long sequence alignments to study the evolution and regulation of mammalian globin gene clusters.
Hardison R, Miller W. Hardison R, et al. Mol Biol Evol. 1993 Jan;10(1):73-102. doi: 10.1093/oxfordjournals.molbev.a039991. Mol Biol Evol. 1993. PMID: 8383794 Review. - Eukaryotic DNA methylation and gene expression.
Weissbach A, Ward C, Bolden A. Weissbach A, et al. Curr Top Cell Regul. 1989;30:1-21. doi: 10.1016/b978-0-12-152830-0.50003-7. Curr Top Cell Regul. 1989. PMID: 2695289 Review. No abstract available.
Cited by
- Targeted microRNA expression in dairy cattle directs production of β-lactoglobulin-free, high-casein milk.
Jabed A, Wagner S, McCracken J, Wells DN, Laible G. Jabed A, et al. Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):16811-6. doi: 10.1073/pnas.1210057109. Epub 2012 Oct 1. Proc Natl Acad Sci U S A. 2012. PMID: 23027958 Free PMC article. - Strategies to insulate lentiviral vector-expressed transgenes.
Ramezani A, Hawley RG. Ramezani A, et al. Methods Mol Biol. 2010;614:77-100. doi: 10.1007/978-1-60761-533-0_5. Methods Mol Biol. 2010. PMID: 20225037 Free PMC article. - Human beta-globin locus control region HS5 contains CTCF- and developmental stage-dependent enhancer-blocking activity in erythroid cells.
Tanimoto K, Sugiura A, Omori A, Felsenfeld G, Engel JD, Fukamizu A. Tanimoto K, et al. Mol Cell Biol. 2003 Dec;23(24):8946-52. doi: 10.1128/MCB.23.24.8946-8952.2003. Mol Cell Biol. 2003. PMID: 14645507 Free PMC article. - Conserved CTCF insulator elements flank the mouse and human beta-globin loci.
Farrell CM, West AG, Felsenfeld G. Farrell CM, et al. Mol Cell Biol. 2002 Jun;22(11):3820-31. doi: 10.1128/MCB.22.11.3820-3831.2002. Mol Cell Biol. 2002. PMID: 11997516 Free PMC article. - Comparison of insulators and promoters for expression of the Wiskott-Aldrich syndrome protein using lentiviral vectors.
Koldej RM, Carney G, Wielgosz MM, Zhou S, Zhan J, Sorrentino BP, Nienhuis AW. Koldej RM, et al. Hum Gene Ther Clin Dev. 2013 Jun;24(2):77-85. doi: 10.1089/humc.2012.244. Epub 2013 Jun 20. Hum Gene Ther Clin Dev. 2013. PMID: 23786330 Free PMC article.
References
- Eissenberg J C, Elgin S C R. Trends Genet. 1991;7:335–340. - PubMed
- Kellum R, Schedl P. Cell. 1991;64:941–950. - PubMed
- Chung J H, Whitely M, Felsenfeld G. Cell. 1993;74:505–514. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases