Rapid determination of single base mismatch mutations in DNA hybrids by direct electric field control - PubMed (original) (raw)
Rapid determination of single base mismatch mutations in DNA hybrids by direct electric field control
R G Sosnowski et al. Proc Natl Acad Sci U S A. 1997.
Abstract
We have demonstrated that controlled electric fields can be used to regulate transport, concentration, hybridization, and denaturation of single- and double-stranded oligonucleotides. Discrimination among oligonucleotide hybrids with widely varying binding strengths may be attained by simple adjustment of the electric field strength. When this approach is used, electric field denaturation control allows single base pair mismatch discrimination to be carried out rapidly (<15 sec) and with high resolution. Electric field denaturation takes place at temperatures well below the melting point of the hybrids, and it may constitute a novel mechanism of DNA denaturation.
Figures
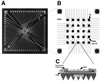
Figure 1
Silicon chip with an array of electrodes. (A) Overview of the microfabricated device. Chip dimension was 1 cm square. Light squares along the perimeter are exposed platinum contact electrodes for connections to power supply. Light lines are platinum leads insulated with dielectric, connecting contact electrodes to the exposed platinum electrodes. (B) Electrode array region of the chip. The central 1 × 1 mm test site array region consists of 4 large (160-μm diameter) corner electrodes and 25 central 80-μm electrodes. Pt, exposed Pt test sites; Si3N4, dielectric; Si3N4 over Pt, Pt insulated by Si3N4. (C) Cross section of an electrode test site. Location of section is indicated by lines extending from B. Pt, Si3N4, Pt insulated by Si3N4; SiO2, dielectric layer; Si substrate, wafer material; Permeation Layer, agarose layer containing streptavidin; DNA, biotinylated oligonucleotides bound to streptavidin. DNA binding was not limited to the surface.
Figure 2
Localization, concentration, and biotin-streptavidin-mediated immobilization of a charged molecule in an electric field. Chips were prepared as described in the legend of Fig. 1 and in the text. A dc electric field was established on the buffer-equilibrated (250 mM cysteine, pH 5.2) chip with C1 as the positive electrode and C2 as the negative electrode. C3 was neutral. Current applied was 100 nA. The buffer was then removed and replaced with a cysteine buffer solution containing 25 nM biotinylated oligonucleotide (T12) conjugated to a Bodipy Texas red fluorophore. To demonstrate immobilization, after 15 sec, two large corner electrodes (Fig. 1) were switched positive and the remainder of the electrodes were switched negative. The current was maintained for 1 min with cysteine buffer washes. The image was captured after washes. R, row; C, column; R3, biotinylated, Bodipy Texas red-conjugated T12; R5, nonbiotinylated, Bodipy Texas red-conjugated T12.
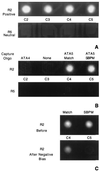
Figure 3
Sequence-specific hybridization and SBPM discrimination facilitated by an electric field. (A) Addressing and concentration of DNA. Biotinylated nonfluorescent capture oligonucleotides were addressed to different test sites as described in the legend of Fig. 2. Row 2 and row 5 were identical: C2, ATA4; C3, none; C4, ATA5 match; C5, ATA5 SBPM. After washing, C2–5 of row 2 were switched positive and the four large corner electrodes were switched negative. Row 5, C2–5 were kept neutral. The current was 100 nA per positive electrode. A solution containing 25 nM Bodipy Texas red-conjugated RCA5 (the complement of ATA5) was applied in 250 mM cysteine buffer. Images were acquired after a 10-sec accumulation of fluorescent DNA over positively biased electrodes. The background is high because of fluorescent DNA in the solution. (B) Sequence-specific hybridization. After 15 sec, NaCl was added to the chip to a final concentration of 250 mM. The electric field was then immediately reversed as described for Fig. 2. Two fluidic washes were performed with 20 mM sodium phosphate, pH 7.4. Images were acquired of the fluorescence remaining on the electrodes after washing. (C) SBPM discrimination. The chip used in A and B was washed five times with 15 μl of 20 mM sodium phosphate, pH 7.0. The four large corner electrodes were switched positive and array electrode R2, C4 was switched negative. A pulsed electric current of 0.6 μA, 0.1 sec on, 0.2 sec off, for 150 cycles was applied through the electrode. This was then repeated at electrode R2, C5. Before, image of electrodes with RCA5 match and mismatch hybrids before electric field denaturation. After Negative Bias, image of the electrodes containing RCA5 match and mismatch hybrids after the electric field pulse sweep had been individually applied at each test site.
Figure 4
Discrimination of match from SBPM for genomically different oligonucleotide hybrids on the same chip. ATA5 match and SBPM capture oligonucleotides were addressed to all five electrodes of C1 and C2, respectively. Ras match and SBPM capture oligonucleotides were directed to C4 and C5, respectively. RCA5 and Ras reporter oligonucleotides were passively hybridized to the previously addressed capture oligonucleotides. The hybridization conditions were 1 μM for each oligonucleotide in 50 mM sodium phosphate/500 mM NaCl, pH 7.4, in 15 μl at room temperature for 5 min. Electric field denaturations were done after equilibration in 20 mM sodium phosphate buffer, as described in the legend of Fig. 3 and below. Images were captured at 1-sec intervals, and average pixel intensity (API) was determined over the electrode area. Data were normalized by dividing the API at each time point by the API at t = 0, × 100. Points represent the average percent fluorescence remaining on 15 replicate test sites, from three separate chips in three separate experiments. Error bars represent the standard deviation at each point. The electric field sweeps took approximately 45 sec. (A) Test sites containing either RCA5 Match or RCA5 SBPM sequence (19-mers) were subjected to current pulses of 0.6 μA, 0.1 sec on, 0.2 sec off, 150 cycles. (B) Test sites containing either Ras Match or Ras SBPM (22-mers) received current pulses of 1.5 μA, 0.1 sec on, 0.2 sec off, 150 cycles.
Similar articles
- Electrostatic surface plasmon resonance: direct electric field-induced hybridization and denaturation in monolayer nucleic acid films and label-free discrimination of base mismatches.
Heaton RJ, Peterson AW, Georgiadis RM. Heaton RJ, et al. Proc Natl Acad Sci U S A. 2001 Mar 27;98(7):3701-4. doi: 10.1073/pnas.071623998. Epub 2001 Mar 20. Proc Natl Acad Sci U S A. 2001. PMID: 11259682 Free PMC article. - Detection of point mutation and insertion mutations in DNA using a quartz crystal microbalance and MutS, a mismatch binding protein.
Su X, Robelek R, Wu Y, Wang G, Knoll W. Su X, et al. Anal Chem. 2004 Jan 15;76(2):489-94. doi: 10.1021/ac035175g. Anal Chem. 2004. PMID: 14719903 - Supramolecular DNA-streptavidin nanocircles with a covalently attached oligonucleotide moiety.
Niemeyer CM, Adler M, Gao S, Chi L. Niemeyer CM, et al. J Biomol Struct Dyn. 2002 Oct;20(2):223-30. doi: 10.1080/07391102.2002.10506838. J Biomol Struct Dyn. 2002. PMID: 12354074 - Improved DNA hybridization parameters by Twisted Intercalating Nucleic Acid (TINA).
Schneider UV. Schneider UV. Dan Med J. 2012 Jan;59(1):B4377. Dan Med J. 2012. PMID: 22239845 Review. - DNA chips: an array of possibilities.
Marshall A, Hodgson J. Marshall A, et al. Nat Biotechnol. 1998 Jan;16(1):27-31. doi: 10.1038/nbt0198-27. Nat Biotechnol. 1998. PMID: 9447589 Review. No abstract available.
Cited by
- Rapid, high fidelity analysis of simple sequence repeats on an electronically active DNA microchip.
Radtkey R, Feng L, Muralhidar M, Duhon M, Canter D, DiPierro D, Fallon S, Tu E, McElfresh K, Nerenberg M, Sosnowski R. Radtkey R, et al. Nucleic Acids Res. 2000 Apr 1;28(7):E17. doi: 10.1093/nar/28.7.e17. Nucleic Acids Res. 2000. PMID: 10710434 Free PMC article. - Fractionation, phosphorylation and ligation on oligonucleotide microchips to enhance sequencing by hybridization.
Dubiley S, Kirillov E, Lysov Y, Mirzabekov A. Dubiley S, et al. Nucleic Acids Res. 1997 Jun 15;25(12):2259-65. doi: 10.1093/nar/25.12.2259. Nucleic Acids Res. 1997. PMID: 9171075 Free PMC article. - SNP analysis using CataCleave probes.
Harvey JJ, Brant SR, Knutson JR, Han MK. Harvey JJ, et al. J Clin Lab Anal. 2008;22(3):192-203. doi: 10.1002/jcla.20240. J Clin Lab Anal. 2008. PMID: 18484652 Free PMC article. - Conformational Changes of Immobilized Polythymine due to External Stressors Studied with Temperature-Controlled Electrochemical Microdevices.
Vishnubhotla R, Montgomery CB, Steffens KL, Semancik S. Vishnubhotla R, et al. Langmuir. 2021 Mar 2;37(8):2607-2618. doi: 10.1021/acs.langmuir.0c03219. Epub 2021 Feb 17. Langmuir. 2021. PMID: 33595321 Free PMC article. - Electric field directed assembly of high-density microbead arrays.
Barbee KD, Hsiao AP, Heller MJ, Huang X. Barbee KD, et al. Lab Chip. 2009 Nov 21;9(22):3268-74. doi: 10.1039/b912876j. Epub 2009 Sep 15. Lab Chip. 2009. PMID: 19865735 Free PMC article.
References
- Fodor S P A, Read J L, Pirrung M C, Stryer L, Lu A T, Solas D. Science. 1991;251:767–773. - PubMed
- Fodor S P, Rava R P, Huang X C, Pease A C, Holmes C P, Adams C L. Nature (London) 1993;364:555–556. - PubMed
- Eggers M, Hogan M, Reich R K, Lamture J, Ehrlich D, Hollis M, Kosicki B, Powdrill T, Beattie K, Smith S, Varma R, Gangadharan R, Mallik A, Burke B, Wallace D. Biotechniques. 1994;17:516–519. - PubMed
- Bains W, Smith G C. J Theor Biol. 1988;135:303–307. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources