Transcriptional regulation in Archaea: in vivo demonstration of a repressor binding site in a methanogen - PubMed (original) (raw)
Transcriptional regulation in Archaea: in vivo demonstration of a repressor binding site in a methanogen
R Cohen-Kupiec et al. Proc Natl Acad Sci U S A. 1997.
Abstract
The status of the Archaea as one of the three primary Domains emphasizes the importance of understanding their molecular fundamentals. Basic transcription in the Archaea resembles eucaryal transcription. However, little is known about transcriptional regulation. We have taken an in vivo approach, using genetics to address transcriptional regulation in the methanogenic Archaeon Methanococcus maripaludis. We identified a repressor binding site that regulates nif (nitrogen fixation) gene expression. The repressor binding site was palindromic (an inverted repeat) and was located just after the transcription start site of nifH. Mutations that changed the sequence of the palindrome resulted in marked decreases in repression by ammonia, even when the palindromic nature of the site was retained. The same mutations greatly decreased binding to the site by components of cell extract. These results provide the first partial description of a transcriptional regulatory mechanism in the methanogenic Archaea. This work also illustrates the utility of genetic approaches in Methanococcus that have not been widely used in the methanogens: directed mutagenesis and reporter gene fusions with lacZ.
Figures
Figure 1
Plasmids for introducing lacZ driven by various promoters into M. maripaludis. The X before a restriction enzyme indicates a site that was removed. Pmcr and Tmcr indicate the methylreductase promoter and terminator, PurR indicates the puromycin resistance gene, and PnifH indicates the nifH promoter.
Figure 2
Nucleotide sequence of nifH promoter region and mutations. The promoter and translational start sites are underlined, palindromic sequences overlined, and transcriptional start site indicated by Γ. Mutations are indicated in parentheses. Mutations altering the first, second, and both halves of the first palindrome are designated CT1, AG1, and CT1AG1, respectively. Similar mutations in the second palindrome are CT2, AG2, and CT2AG2. A mutation making the first palindrome identical to the second is TA3T, and a mutation deleting six nucleotides between the two palindromes is Δ6.
Figure 3
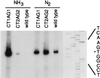
Primer extension analysis of the nifH promoter–lacZ transcript produced from wild-type or mutant promoter regions during growth on ammonia or N2. Visualization of total RNA after gel electrophoresis indicated that the quantity of RNA was similar for all samples.
Figure 4
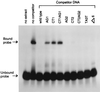
Electrophoretic mobility-shift assay for binding of M. maripaludis extract to wild-type and mutant palindrome-containing region. A 1000-fold excess of unlabeled competitor DNA was tested for its ability to prevent the mobility shift caused by binding of cell extract to the labeled wild-type region.
Similar articles
- Genetics of nitrogen regulation in Methanococcus maripaludis.
Kessler PS, Leigh JA. Kessler PS, et al. Genetics. 1999 Aug;152(4):1343-51. doi: 10.1093/genetics/152.4.1343. Genetics. 1999. PMID: 10430565 Free PMC article. - The nif gene operon of the methanogenic archaeon Methanococcus maripaludis.
Kessler PS, Blank C, Leigh JA. Kessler PS, et al. J Bacteriol. 1998 Mar;180(6):1504-11. doi: 10.1128/JB.180.6.1504-1511.1998. J Bacteriol. 1998. PMID: 9515920 Free PMC article. - Regulatory response of Methanococcus maripaludis to alanine, an intermediate nitrogen source.
Lie TJ, Leigh JA. Lie TJ, et al. J Bacteriol. 2002 Oct;184(19):5301-6. doi: 10.1128/JB.184.19.5301-5306.2002. J Bacteriol. 2002. PMID: 12218015 Free PMC article. - Nitrogen fixation in methanogens: the archaeal perspective.
Leigh JA. Leigh JA. Curr Issues Mol Biol. 2000 Oct;2(4):125-31. Curr Issues Mol Biol. 2000. PMID: 11471757 Review. - Genetics of Methanococcus: possibilities for functional genomics in Archaea.
Tumbula DL, Whitman WB. Tumbula DL, et al. Mol Microbiol. 1999 Jul;33(1):1-7. doi: 10.1046/j.1365-2958.1999.01463.x. Mol Microbiol. 1999. PMID: 10411718 Review.
Cited by
- Expression vectors for Methanococcus maripaludis: overexpression of acetohydroxyacid synthase and beta-galactosidase.
Gardner WL, Whitman WB. Gardner WL, et al. Genetics. 1999 Aug;152(4):1439-47. doi: 10.1093/genetics/152.4.1439. Genetics. 1999. PMID: 10430574 Free PMC article. - Function and regulation of the formate dehydrogenase genes of the methanogenic archaeon Methanococcus maripaludis.
Wood GE, Haydock AK, Leigh JA. Wood GE, et al. J Bacteriol. 2003 Apr;185(8):2548-54. doi: 10.1128/JB.185.8.2548-2554.2003. J Bacteriol. 2003. PMID: 12670979 Free PMC article. - Genetics of nitrogen regulation in Methanococcus maripaludis.
Kessler PS, Leigh JA. Kessler PS, et al. Genetics. 1999 Aug;152(4):1343-51. doi: 10.1093/genetics/152.4.1343. Genetics. 1999. PMID: 10430565 Free PMC article. - Characterization of GlnK1 from Methanosarcina mazei strain Gö1: complementation of an Escherichia coli glnK mutant strain by GlnK1.
Ehlers C, Grabbe R, Veit K, Schmitz RA. Ehlers C, et al. J Bacteriol. 2002 Feb;184(4):1028-40. doi: 10.1128/jb.184.4.1028-1040.2002. J Bacteriol. 2002. PMID: 11807063 Free PMC article.
References
- Baumann P, Qureshi S A, Jackson S P. Trends Genet. 1995;11:279–283. - PubMed
- Rowlands T, Baumann P, Jackson S P. Science. 1994;264:1326–1329. - PubMed
- Ouzounis C, Sander C. Cell. 1992;71:189–190. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources