Interconversion of red opsin isoforms by the cyclophilin-related chaperone protein Ran-binding protein 2 - PubMed (original) (raw)
Interconversion of red opsin isoforms by the cyclophilin-related chaperone protein Ran-binding protein 2
P A Ferreira et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Ran-binding protein 2 (RanBP2) (type II) is a retinal cyclophilin-related protein that binds Ran-GTPase. Type I cyclophilin is a shorter, alternatively spliced isoform of RanBP2. Recently, we showed that the Ran-binding domain 4 (RBD4)/cyclophilin (CY) supradomain of RanBP2 acts both in vitro and in vivo as a specific chaperone for bovine red/green opsin (R/G opsin). R/G opsin undergoes a stable modification of its electrophoretic mobility upon binding to RanBP2. This modification is likely due to cis-trans isomerization of one or more proline residues in the opsin protein. Here, we show that expression of human red opsin in Escherichia coli and COS cells results in the production of still a third electrophoretic variant of this protein. This variant was converted to the RBD4 binding-competent form of opsin through direct interaction with RBD4/CY, both in vivo and in vitro. We suggest that these distinct opsin species may represent kinetically or thermodynamically trapped prolyl conformers that can be interconverted by concerted action of the RBD4 and CY domains of RanBP2. We also show that the C-terminal half of RBD4 is the binding domain for bovine R/G opsin and that coexpression of human red opsin with type I cyclophilin in vivo enhances the production of functional visual pigment. These observations imply that prolyl isomerization may have importance beyond its role in protein folding, possibly as a molecular switch modulated by cyclophilin for the loading of opsin onto RanBP2 during visual pigment processing in cones.
Figures
Figure 1
Schematic diagram of six bovine RBD4/CY recombinant constructs used in this study. RBD4/CY contains the entire RBD4 and CY with PPIase catalytic site. RBD4-C/CY contains the C-terminal half of RBD4 and the whole CY. This construct represents type I cyclophilin, assuming the second methionine is used for translation initiation (33). CY contains only the cyclophilin domain. RBD4 contains the entire RBD4. N-RBD4 contains the N-terminal half of RBD4. RBD4-C contains the C-terminal half of RBD4. The restriction sites of the cognate cDNA used for the cloning of the RanBP2 domains into the GST-expression vector, pGEX-KG, are shown below the constructs. B, _Bam_HI; Bst, _Bst_XI; N, _Nco_I; S, _Stu_I.
Figure 2
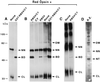
The C-terminal half of RBD4 is the binding domain for bovine R/G opsin. Western blot analysis of glutathione-_S_-agarose coprecipitates from incubation reactions of bovine retinal extracts with GST-RBD4/CY and subfragments thereof (Fig. 1) at 4°C (Left) and 26°C (Right) using an antibody against human red opsin (41). Removal of the N-terminal half of RBD4 from RBD4/CY (RBD4-C/CY) abolishes its binding to R/G opsin at 4°C. This binding could be restored by further removal of CY from RBD4-C/CY (RBD4-C). The increase of the incubation temperature from 4°C to 26°C made RBD4-C/CY competent to bind R/G opsin without changing the affinity of R/G opsin to any other constructs. CL, NT, BD, and DM represent, respectively, the 34-kDa collapsed, traces of the 49.5-kDa native, 51-kDa RBD4/CY-binding competent form, and traces of red opsin dimers.
Figure 3
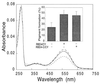
UV–visible absorption spectra of red pigment expressed in COS cells. Red opsin expressed with RBD4-C/CY (type I cyclophilin) (solid line), with RBD4-CY (dotted line), and alone (broken line) was reconstituted with 11-_cis_-retinal and purified as described in the text. Coexpression of red opsin with type I cyclophilin [the second starting methionine (33) was used as translational start site] increased pigment generation similar to that as with RBD4-CY (Inset). Similar results were obtained when the first starting methionine of type I cyclophilin was used as a translational start site (data not shown). The spectra were normalized to the same scale at 280 nm absorbance. The typical absolute 280 values ranged from 0.07 to 0.8 for red opsin alone, from 0.6 to 0.8 for red opsin with type I cyclophilin, and from 0.5 to 0.7 for red opsin with RBD4-CY.
Figure 4
In vivo and in vitro binding and electrophoretic mobility-shift assays of RBD4/CY, RBD4, and CY on red opsin. (A) Western blot of red opsin expressed by itself or coexpressed with GST-RBD4/CY in E. coli using an antibody against red opsin. (B) Western blot of purified red opsin from E. coli by itself and incubated with purified RBD4/CY, RBD4, and CY using an antibody against red opsin. Lanes 4 and 5 represent incubations with opsin purified from E. coli in the absence of detergent. (C) Western blot of red opsin purified from COS cells by itself and incubated with purified RBD4/CY. (D) Western blot of an aliquot of crude retinal extracts with an antibody against red opsin. A major 65-kDa opsin isoform was produced when red opsin was expressed either in E. coli or in COS cells. This opsin isoform may be converted to the 51-kDa RBD4/CY-binding-competent form by the concerted action of RBD4 and CY. The 65-kDa opsin isoform is a very minor species in retinal extracts, while the RBD4/CY-binding-competent form of opsin (51 kDa) is undetectable. In contrast, the 49.5-kDa (native) isoform is the major opsin isoform in retinal extracts. The 86-kDa and 34-kDa bands may represent SDS/PAGE-resistant opsin dimers and collapsed isoforms of red opsin, respectively. High molecular weight SDS/PAGE-resistant aggregates of opsin were seen in retinal extracts (D) and with opsin expressed in COS cells (C). All samples were run on the same SDS/polyacrylamide gel. CL, NT, BD, NN, and DM represent, respectively, the 34-kDa collapsed, 49.5-kDa native, 51-kDa RBD4/CY-binding-competent form, 65-kDa nonnative, and dimers of red opsin. R.E., bovine retinal extract.
Figure 5
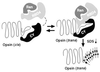
Schematic model of the interaction between the RBD4/CY supradomain and opsin. In the absence of added RBD4/CY, the majority of R/G opsin is in the _cis_-isomerized form. The prolyl-foldase, CY, induces cis to trans isomerization of one or more peptidyl-prolyl bonds in opsin, resulting in a form that is competent to bind RBD4. CY also is required as a chaperone to stabilize the interaction between opsin and RBD4. SDS stabilizes the trans form of opsin, which has a low propensity for self-aggregation. The cellular role of Ran-GTPase in opsin folding/processing is still unclear. It may be involved in the nuclear export of opsin mRNA, and with RBD4, may act as a CY-mediated coupling factor between transcription and translation of opsin(s) (38).
Similar articles
- Cyclophilin-related protein RanBP2 acts as chaperone for red/green opsin.
Ferreira PA, Nakayama TA, Pak WL, Travis GH. Ferreira PA, et al. Nature. 1996 Oct 17;383(6601):637-40. doi: 10.1038/383637a0. Nature. 1996. PMID: 8857542 - Differential loss of prolyl isomerase or chaperone activity of Ran-binding protein 2 (Ranbp2) unveils distinct physiological roles of its cyclophilin domain in proteostasis.
Cho KI, Patil H, Senda E, Wang J, Yi H, Qiu S, Yoon D, Yu M, Orry A, Peachey NS, Ferreira PA. Cho KI, et al. J Biol Chem. 2014 Feb 21;289(8):4600-25. doi: 10.1074/jbc.M113.538215. Epub 2014 Jan 8. J Biol Chem. 2014. PMID: 24403063 Free PMC article. - Targeting the cyclophilin domain of Ran-binding protein 2 (Ranbp2) with novel small molecules to control the proteostasis of STAT3, hnRNPA2B1 and M-opsin.
Cho KI, Orry A, Park SE, Ferreira PA. Cho KI, et al. ACS Chem Neurosci. 2015 Aug 19;6(8):1476-85. doi: 10.1021/acschemneuro.5b00134. Epub 2015 Jun 12. ACS Chem Neurosci. 2015. PMID: 26030368 Free PMC article. - Mechanism of enzymatic and nonenzymatic prolyl cis-trans isomerization.
Stein RL. Stein RL. Adv Protein Chem. 1993;44:1-24. doi: 10.1016/s0065-3233(08)60562-8. Adv Protein Chem. 1993. PMID: 8317295 Review. No abstract available. - The Escherichia coli trigger factor.
Hesterkamp T, Bukau B. Hesterkamp T, et al. FEBS Lett. 1996 Jun 24;389(1):32-4. doi: 10.1016/0014-5793(96)00582-0. FEBS Lett. 1996. PMID: 8682200 Review.
Cited by
- Roles of Nucleoporin RanBP2/Nup358 in Acute Necrotizing Encephalopathy Type 1 (ANE1) and Viral Infection.
Jiang J, Wang YE, Palazzo AF, Shen Q. Jiang J, et al. Int J Mol Sci. 2022 Mar 24;23(7):3548. doi: 10.3390/ijms23073548. Int J Mol Sci. 2022. PMID: 35408907 Free PMC article. Review. - Kinesin-1 and mitochondrial motility control by discrimination of structurally equivalent but distinct subdomains in Ran-GTP-binding domains of Ran-binding protein 2.
Patil H, Cho KI, Lee J, Yang Y, Orry A, Ferreira PA. Patil H, et al. Open Biol. 2013 Mar 27;3(3):120183. doi: 10.1098/rsob.120183. Open Biol. 2013. PMID: 23536549 Free PMC article. - Neuroprotection resulting from insufficiency of RANBP2 is associated with the modulation of protein and lipid homeostasis of functionally diverse but linked pathways in response to oxidative stress.
Cho KI, Yi H, Tserentsoodol N, Searle K, Ferreira PA. Cho KI, et al. Dis Model Mech. 2010 Sep-Oct;3(9-10):595-604. doi: 10.1242/dmm.004648. Epub 2010 Aug 3. Dis Model Mech. 2010. PMID: 20682751 Free PMC article. - RanBP2/Nup358 Mediates Sumoylation of STAT1 and Antagonizes Interferon-α-Mediated Antiviral Innate Immunity.
Li J, Su L, Jiang J, Wang YE, Ling Y, Qiu Y, Yu H, Huang Y, Wu J, Jiang S, Zhang T, Palazzo AF, Shen Q. Li J, et al. Int J Mol Sci. 2023 Dec 25;25(1):299. doi: 10.3390/ijms25010299. Int J Mol Sci. 2023. PMID: 38203469 Free PMC article. - Loss of Ranbp2 in motoneurons causes disruption of nucleocytoplasmic and chemokine signaling, proteostasis of hnRNPH3 and Mmp28, and development of amyotrophic lateral sclerosis-like syndromes.
Cho KI, Yoon D, Qiu S, Danziger Z, Grill WM, Wetsel WC, Ferreira PA. Cho KI, et al. Dis Model Mech. 2017 May 1;10(5):559-579. doi: 10.1242/dmm.027730. Epub 2017 Jan 18. Dis Model Mech. 2017. PMID: 28100513 Free PMC article.
References
- Schmid F. Annu Rev Biophys Biomol Struct. 1993;22:123–143. - PubMed
- Fischer G. Angew Chem Int Ed Engl. 1994;33:1415–1436.
- Fischer G. Angew Chem. 1994;106:1479–1501.
- Fischer G. Angew Chem Int Ed Engl. 1994;63:67–118.
- Rudd K, Sophia H, Koonin E, Plunkett G, Lazar S, Rouviere P. Trends Biochem Sci. 1995;20:12–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous