Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells - PubMed (original) (raw)
Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells
N Belgareh et al. J Cell Biol. 1997.
Abstract
To follow the dynamics of nuclear pore distribution in living yeast cells, we have generated fusion proteins between the green fluorescent protein (GFP) and the yeast nucleoporins Nup49p and Nup133p. In nup133- dividing cells that display a constitutive nuclear pore clustering, in vivo analysis of GFP-Nup49p localization revealed changes in the distribution of nuclear pore complex (NPC) clusters. Furthermore, upon induction of Nup133p expression in a GAL-nup133 strain, a progressive fragmentation of the NPC aggregates was observed that in turn led to a wild-type nuclear pore distribution. To try to uncouple Nup133p-induced NPC redistribution from successive nuclear divisions and nuclear pore biogenesis, we devised an assay based on the formation of heterokaryons between nup133- mutants and cells either expressing or overexpressing Nup133p. Under these conditions, the use of GFP-Nup133p and GFP-Nup49p fusion proteins revealed that Nup133p can be rapidly targeted to the clustered nuclear pores, where its amino-terminal domain is required to promote the redistribution of preexisting NPCs.
Figures
Figure 1
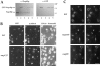
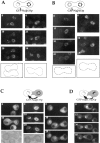
Expression and localization of Nup49p tagged with GFP. (A) Immunoblotting analysis. Whole cell extracts were prepared from yeast strains either containing or not containing the pUN100-GFP-NUP49 plasmid: nup133− (lane 1), nup133− + pUN100-GFPNUP49 (lane 2), VD1R (lane 3), VD1R + pUN100-GFP-NUP49 (lane 4), and VD1R + pUN100-GFP-NUP49 and lacking pCH1122NUP49 (lane 5). Equivalent amounts of extracts from each strain were analyzed by SDS-PAGE and immunoblotting using an antiNup49p and an anti-GFP antibody. The positions of Nup49p and GFP-Nup49p and the molecular masses (kD) of a protein standard are indicated. (B) Colocalization of GFP-Nup49p and nuclear pore complex antigens. Wild-type (WT) and nup133− cells expressing GFPNup49p, which is detected in the fluorescein channel (GFP), were fixed and processed for indirect immunofluorescence using the monoclonal antibody mAb414 followed by rhodamine-labeled goat–anti mouse IgG. DNA staining and the Nomarski optics view are shown. (C) In vivo localization of GFP-Nup49p using confocal microscopy. Two successive sections (0.5 μm apart) are presented. A ringlike staining of the nuclear periphery, typical for nucleoporins, is observed in wild-type cells (WT). Arrows show absence of nuclear pore complexes in areas of close association between the nucleus and the vacuole. In nup133− and to a lesser extent nup85− strains, nuclear pores are clustered. Bars, 5 μm.
Figure 2
Nuclear pore distribution in wild-type and nup133− cells at 10-min intervals. Cells expressing GFP-Nup49p were examined by confocal microscopy as described in Materials and Methods. Each image is a two-dimensional projection of images collected at all relevant z-axes. (A) In wild-type RS453
a
cells, elongation of the nucleus towards the daughter cell can be observed. (B) In synchronized nup133−
a
cells, a dynamic distribution of the clusters of NPCs is observed (left). Soon after cell division (right), both mother and daughter nuclei move towards the future buds; note the position of clustered NPCs close to each novel budneck. During karyokinesis (bottom), elongated clusters that span the two nuclei can also be seen.
Figure 3
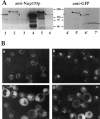
Induction of Nup133p expression in GAL-nup133 cells. (A) Growth properties of GAL-nup133 cells. Two equivalent dilutions of GAL-nup133 cells were spotted on glucose (YPDA)– or galactose (YPGalA)–containing plates and incubated for 3 d at 24°C or 2 d at 37°C. (B) Analysis of Nup133p expression levels by immunoblotting. Whole cell extracts were prepared from the following strains: nup133− (−), wild-type RS453 (+), nup133− carrying the high copy number plasmid pRS424-NUP133 (++), and GAL-nup133 grown overnight in YPDA medium (0) or shifted for 2, 4, 6, 8, and 24 h to YPGalA medium. Total protein corresponding to similar amounts of cells were analyzed by SDS-PAGE and immunoblotting using an anti-Nup133p antibody. AntiNop1p antibody was used to control the amounts of protein loaded. The molecular masses (kD) of a protein standard are indicated. (C) Analysis of NPC distribution during Nup133p induction. Cells from strain GAL-nup133 carrying plasmid pUN100GFP-NUP49 were grown in SDC-leu medium (0) or shifted to SGalC-leu medium for 2, 4, 6, 8, or 24 h as indicated and analyzed by confocal microscopy. Each image is a two-dimensional projection of images collected at all relevant z-axes using the fluorescein channel. The fluorescent staining patterns reflect the intracellular distribution of NPCs.
Figure 4
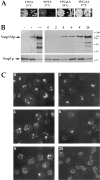
Expression and localization of GFP-Nup133p and GFP-nup133ΔNp. (A) Immunoblotting analysis. To follow the expression of wild-type and GFP-tagged Nup133p, whole cell extracts were analyzed by SDS-PAGE and immunoblotting using either an antibody directed against the amino-terminal domain of Nup133p or an anti-GFP antibody. The following strains were analyzed: kar1-1 (lane 1), nup133− + pUN100-GFP-NUP133 (lane 2), nup133− + pUN100-GFP-nup133ΔN (lane 3), nup133− + pRS424-NUP133 (lanes 4 and 4′), nup133− + pRS424-GFPNUP133 (lanes 5 and 5′), and nup133− + pRS424-GFPnup133ΔN (lanes 6 and 6′). As reference, a whole cell extract from strain VD1R + pUN100-GFP-NUP49 was loaded on the same gel (7′). The positions of Nup133p (○), GFP-Nup133p (•), GFP-nup133ΔNp (Δ), and GFP-Nup49p (*) are indicated. (B) Confocal analysis of nup133− cells expressing GFP-Nup133p and GFP-nup133ΔNp chimeras. (a) Expression of GFP-Nup133p carried by plasmid pUN100 gives rise to a faint, ringlike staining. (b) GFP-nup133ΔNp expressed on the pUN100 centromeric plasmid localizes within NPC clusters. (c and d) When carried by the high copy number plasmid pRS424, GFP-Nup133p (c) and GFPnup133ΔNp (d) present a heterogeneous expression level; some cells display an additional cytoplasmic and nuclear staining. Images correspond to individual sections recorded by confocal microscopy. To enable quantitative comparison, the power intensity of the laser was set to the same value in each case.
Figure 5
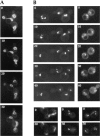
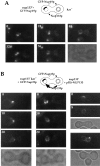
Analysis of GFPNup133p targeting to nup133− and wild-type cells upon mating. (A and B) nup133−α (A) or wild-type (B) cells were mated with nup133−kar1-1
a
cells carrying the pUN100-GFPNUP133 plasmid as described in Materials and Methods. Heterokaryons defective for nuclear fusion were followed by confocal microscopy. Schematics of the heterokaryon boundaries are shown for reference. (C and D) nup133−kar1-1
α
cells were mated with nup133−
a
cells carrying the pRS424-GFP-NUP133 (C) or the pRS424-GFP-nup133ΔN plasmid (D). Heterokaryons were followed by confocal microscopy. Time 0 corresponds to the last image recorded before cytoduction, i.e., when no cytoplasmic staining is detectable in the left (nup133−kar1-1) cells. A transmission image of the first two heterokaryons is shown for reference.
Figure 6
Overexpression of Nup133p increases the rate of NPC redistribution during the mating process. (A) nup133−
α
cells carrying the pUN100-GFP-NUP49 plasmid were mixed with kar1-1
a
cells. An example of such a heterokaryon, followed at 12-min intervals using confocal microscopy, is presented. The transmission image (bottom) shows the morphology of the zygote at the end of the experiment (48 min). (B) nup133−kar1-1
a
cells expressing GFP-Nup49p were mixed with nup133−
α
cells expressing Nup133p carried by the high copy number plasmid pRS424-NUP133. Heterokaryons were followed at 10-min intervals. At time 0 (when the septum between the mating cells was still detectable), the clustered nuclear pores from the nup133−kar1-1 nuclei are stained with GFP-Nup49p. After cytoduction, the redistribution of nuclear pores labeled with GFP-Nup49p occurs within 10–20 min. A faint labeling of the other nuclei can be detectable after 20 min. In the upper heterokaryon, redistribution of the GFP-Nup49p labeling had occurred within 8 min (right). Small aggregates randomly distributed around the nuclear envelope are detectable. The other heterokaryon had already reached a later stage in the mating process (as revealed by its trilobed morphology on the corresponding transmission image) and presents a staining of both nuclei.
Figure 7
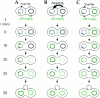
Schematic representation of GFP-Nup49p localization (green) and NPC distribution during the mating process. (A and B) mating between nup133− cells expressing GFP-Nup49p and cells expressing or overexpressing Nup133p (see also Fig. 6). (C and D) Analysis of GFP-Nup49p targeting to the nup133− nucleus upon mating between nup133− cells and cells expressing GFP-Nup49p together with wild-type Nup133p (C) or overexpressed Nup133p (D). The localization of GFP-Nup49p and the shape of heterokaryons recorded on the CCD camera at various stages during the mating process are presented. The delay required for these processes, as estimated through kinetic analysis of individual heterokaryons, is indicated on the left.
Figure 7
Schematic representation of GFP-Nup49p localization (green) and NPC distribution during the mating process. (A and B) mating between nup133− cells expressing GFP-Nup49p and cells expressing or overexpressing Nup133p (see also Fig. 6). (C and D) Analysis of GFP-Nup49p targeting to the nup133− nucleus upon mating between nup133− cells and cells expressing GFP-Nup49p together with wild-type Nup133p (C) or overexpressed Nup133p (D). The localization of GFP-Nup49p and the shape of heterokaryons recorded on the CCD camera at various stages during the mating process are presented. The delay required for these processes, as estimated through kinetic analysis of individual heterokaryons, is indicated on the left.
Figure 7
Schematic representation of GFP-Nup49p localization (green) and NPC distribution during the mating process. (A and B) mating between nup133− cells expressing GFP-Nup49p and cells expressing or overexpressing Nup133p (see also Fig. 6). (C and D) Analysis of GFP-Nup49p targeting to the nup133− nucleus upon mating between nup133− cells and cells expressing GFP-Nup49p together with wild-type Nup133p (C) or overexpressed Nup133p (D). The localization of GFP-Nup49p and the shape of heterokaryons recorded on the CCD camera at various stages during the mating process are presented. The delay required for these processes, as estimated through kinetic analysis of individual heterokaryons, is indicated on the left.
Figure 7
Schematic representation of GFP-Nup49p localization (green) and NPC distribution during the mating process. (A and B) mating between nup133− cells expressing GFP-Nup49p and cells expressing or overexpressing Nup133p (see also Fig. 6). (C and D) Analysis of GFP-Nup49p targeting to the nup133− nucleus upon mating between nup133− cells and cells expressing GFP-Nup49p together with wild-type Nup133p (C) or overexpressed Nup133p (D). The localization of GFP-Nup49p and the shape of heterokaryons recorded on the CCD camera at various stages during the mating process are presented. The delay required for these processes, as estimated through kinetic analysis of individual heterokaryons, is indicated on the left.
Similar articles
- In vivo dynamics of nuclear pore complexes in yeast.
Bucci M, Wente SR. Bucci M, et al. J Cell Biol. 1997 Mar 24;136(6):1185-99. doi: 10.1083/jcb.136.6.1185. J Cell Biol. 1997. PMID: 9087436 Free PMC article. - Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes.
Li O, Heath CV, Amberg DC, Dockendorff TC, Copeland CS, Snyder M, Cole CN. Li O, et al. Mol Biol Cell. 1995 Apr;6(4):401-417. doi: 10.1091/mbc.6.4.401. Mol Biol Cell. 1995. PMID: 7626806 Free PMC article. - Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes.
Pemberton LF, Rout MP, Blobel G. Pemberton LF, et al. Proc Natl Acad Sci U S A. 1995 Feb 14;92(4):1187-91. doi: 10.1073/pnas.92.4.1187. Proc Natl Acad Sci U S A. 1995. PMID: 7862658 Free PMC article. - Genetic approaches to nuclear pore structure and function.
Doye V, Hurt EC. Doye V, et al. Trends Genet. 1995 Jun;11(6):235-41. doi: 10.1016/s0168-9525(00)89057-5. Trends Genet. 1995. PMID: 7638906 Review. - The nuclear pore complex.
Hurt EC. Hurt EC. FEBS Lett. 1993 Jun 28;325(1-2):76-80. doi: 10.1016/0014-5793(93)81417-x. FEBS Lett. 1993. PMID: 8513897 Review.
Cited by
- The Malleable Nature of the Budding Yeast Nuclear Envelope: Flares, Fusion, and Fenestrations.
Meseroll RA, Cohen-Fix O. Meseroll RA, et al. J Cell Physiol. 2016 Nov;231(11):2353-60. doi: 10.1002/jcp.25355. Epub 2016 Apr 8. J Cell Physiol. 2016. PMID: 26909870 Free PMC article. Review. - An essential nuclear envelope integral membrane protein, Brr6p, required for nuclear transport.
de Bruyn Kops A, Guthrie C. de Bruyn Kops A, et al. EMBO J. 2001 Aug 1;20(15):4183-93. doi: 10.1093/emboj/20.15.4183. EMBO J. 2001. PMID: 11483521 Free PMC article. - Immobility, inheritance and plasticity of shape of the yeast nucleus.
Hattier T, Andrulis ED, Tartakoff AM. Hattier T, et al. BMC Cell Biol. 2007 Nov 9;8:47. doi: 10.1186/1471-2121-8-47. BMC Cell Biol. 2007. PMID: 17996101 Free PMC article. - Depletion of the yeast nuclear exosome subunit Rrp6 results in accumulation of polyadenylated RNAs in a discrete domain within the nucleolus.
Carneiro T, Carvalho C, Braga J, Rino J, Milligan L, Tollervey D, Carmo-Fonseca M. Carneiro T, et al. Mol Cell Biol. 2007 Jun;27(11):4157-65. doi: 10.1128/MCB.00120-07. Epub 2007 Apr 2. Mol Cell Biol. 2007. PMID: 17403903 Free PMC article. - The transmission of nuclear pore complexes to daughter cells requires a cytoplasmic pool of Nsp1.
Colombi P, Webster BM, Fröhlich F, Lusk CP. Colombi P, et al. J Cell Biol. 2013 Oct 28;203(2):215-32. doi: 10.1083/jcb.201305115. J Cell Biol. 2013. PMID: 24165936 Free PMC article.
References
- Allen JL, Douglas MG. Organization of the nuclear pore complex in Saccharomyces cerevisiae. . J Ultrastruct Mol Struct Res. 1989;102:95–108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases