Development of "substrate-trapping" mutants to identify physiological substrates of protein tyrosine phosphatases - PubMed (original) (raw)
Development of "substrate-trapping" mutants to identify physiological substrates of protein tyrosine phosphatases
A J Flint et al. Proc Natl Acad Sci U S A. 1997.
Abstract
The identification of substrates of protein tyrosine phosphatases (PTPs) is an essential step toward a complete understanding of the physiological function of members of this enzyme family. PTPs are defined by a conserved catalytic domain harboring 27 invariant residues. From a mutagenesis study of these invariant residues that was guided by our knowledge of the crystal structure of PTP1B, we have discovered a mutation of the invariant catalytic acid (Asp-181 in PTP1B) that converts an extremely active enzyme into a "substrate trap." Expression of this D181A mutant of PTP1B in COS and 293 cells results in an enzyme that competes with endogenous PTP1B for substrates and promotes the accumulation of phosphotyrosine primarily on the epidermal growth factor (EGF) receptor as well as on proteins of 120, 80, and 70 kDa. The association between the D181A mutant of PTP1B and these substrates was sufficiently stable to allow isolation of the complex by immunoprecipitation. As predicted for an interaction between the substrate-binding site of PTP1B and its substrates, the complex is disrupted by vanadate and, for the EGF receptor, the interaction absolutely requires receptor autophosphorylation. Furthermore, from immunofluorescence studies, the D181A mutant of PTP1B appeared to retain the endogenous EGF receptor in an intracellular complex. These results suggest that the EGF receptor is a bona fide substrate for PTP1B in vivo and that one important function of PTP1B is to prevent the inappropriate, ligand-independent, activation of newly synthesized EGF receptor in the endoplasmic reticulum. This essential catalytic aspartate residue is present in all PTPs and has structurally equivalent counterparts in the dual-specificity phosphatases and the low molecular weight PTPs. Therefore we anticipate that this method may be widely applicable to facilitate the identification of substrates of other members of this enzyme family.
Figures
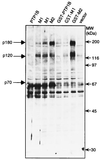
Figure 1
Induction of tyrosine phosphorylation resulting from expression of PTP1B mutants in COS cells. Samples (50 μg) of protein from lysates of COS cells transfected with pMT2 alone (vector) or with pMT2 plasmids expressing wild-type PTP1B, PTP1B with a C215A mutation (M1), PTP1B with a D181A mutation (M2), or GST fusions of each of these proteins were immunoblotted with anti-phosphotyrosine mAb 4G10. Proteins showing substantial increases in their phosphotyrosine content are marked with the dark arrows on the left. The positions of the molecular mass markers (Sigma) myosin, β-galactosidase, phosphorylase, bovine serum albumin, ovalbumin, and carbonic anhydrase are marked on the right with thin arrows.
Figure 2
Precipitation of tyrosine-phosphorylated proteins in association with the D181A (M2) mutant of PTP1B. Immunoprecipitates of PTP1B from lysates of COS cells transfected to express PTP1B, C215A (M1), or D181A (M2) and glutathione-Sepharose precipitates from lysates of COS cells transfected to express GST fusions to each of these proteins were immunoblotted with anti-phosphotyrosine mAb 4G10. Pairs of lanes represent samples from duplicate transfections. A − indicates that for 24 h prior to harvesting the cells were maintained in media without serum. Lanes marked with + indicate that these cells were similarly deprived of serum until 10 min before harvesting, during which time they were incubated with 20% fetal bovine serum, a treatment that did not significantly alter the phosphotyrosine content of proteins associated with D181A-PTP1B. The three intense black bands found in all the anti-PTP1B immunoprecipitates, including the vector only control, are derived from the precipitating mAb. (Right) Anti-pTyr (4G10) blot of 50 μg of lysate from untransfected COS cells that were untreated (none) or incubated for 10 min with EGF at 100 ng/ml or platelet-derived growth factor (PDGF) at 5 ng/ml.
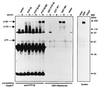
Figure 3
Identification of tyrosine-phosphorylated p180 in the D181A precipitates as the EGFR. This experiment was conducted as described in the legend of Fig. 2 except that the resulting immunoprecipitates were split in half, subjected to electrophoresis on duplicate SDS/polyacrylamide gels, and blotted with either anti-EGFR mAb (Transduction Laboratories) (Upper Left) or with anti-pTyr (4G10) (Lower Left). Pairs of lanes represent samples from duplicate transfections. A − indicates that for 24 h prior to harvesting the cells were maintained in media without serum. A + indicates that these cells were similarly deprived of serum until 10 min before harvesting, during which time they were incubated with EGF at 100 ng/ml. The anti-pTyr blot was deliberately overexposed in an attempt to observe pTyr-containing proteins in the C215A (M1) precipitates and to visualize p70 and p80 in the D181A (M2) precipitates. Anti-EGFR staining was weaker than anti-pTyr staining of p180 in D181A (M2) precipitates, presumably either because the anti-EGFR antibody is of lower affinity or because there are multiple pTyr epitopes recognized by 4G10 in the “trapped” EGFR. (Right) Anti-EGFR blot of 50 μg of lysate from untransfected COS cells that were untreated (−) or incubated for 10 min with EGF at 100 ng/ml (+).
Figure 4
Reconstitution of the interaction between D181A and the EGFR requires EGFR autophosphorylation. 293 cells were cotransfected with plasmids expressing PTP1B, the C215A or D181A PTP1B mutants, and plasmids expressing the human EGFR or a catalytically inactive (K−) mutant, in which the Lys responsible for coordinating ATP, Lys-721, was converted to Arg, or a C-terminally truncated (c′958) form that lacks all the major autophosphorylation sites. Immunoprecipitates of PTP1B were split in half and resolved by SDS/PAGE on duplicate gels. One gel (Upper) was blotted with anti-EGFR antibody KSM and the other (Lower) was blotted for pTyr with G98. Panels on each end represent immunoblots of 50 μg of lysate from the cells cotransfected with D181A-PTP18 (M2) and the various EGFR forms. CMV, cytomegalovirus. Although both the K− and c′958 forms of EGFR are expressed well, neither contains pTyr, and both fail to interact with the D181A mutant of PTP1B.
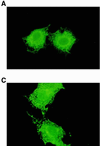
Figure 5
Colocalization of D181A-PTP1B and endogenous EGFRs. COS 1 cells transfected with wild-type PTP1B (data not shown) or C215A (A and B) or D181A expression plasmids (C and D) were fixed with paraformaldehyde at 36 h after transfection and processed for immunofluorescence as previously described (24). Cells were incubated with anti-PTP1B FG6 ascites fluid (10) and anti-EGFR polyclonal serum (1964, kindly provided by G. Gill) at dilutions of 1/4000 and 1/500, respectively. Overexpressed PTP1B (A and C) and endogenous EGFR (B and D) were visualized with fluorescein-conjugated sheep anti-mouse and Texas red-conjugated goat anti-rabbit antibodies (Cappel), respectively.
Figure 5
Colocalization of D181A-PTP1B and endogenous EGFRs. COS 1 cells transfected with wild-type PTP1B (data not shown) or C215A (A and B) or D181A expression plasmids (C and D) were fixed with paraformaldehyde at 36 h after transfection and processed for immunofluorescence as previously described (24). Cells were incubated with anti-PTP1B FG6 ascites fluid (10) and anti-EGFR polyclonal serum (1964, kindly provided by G. Gill) at dilutions of 1/4000 and 1/500, respectively. Overexpressed PTP1B (A and C) and endogenous EGFR (B and D) were visualized with fluorescein-conjugated sheep anti-mouse and Texas red-conjugated goat anti-rabbit antibodies (Cappel), respectively.
Figure 6
The analogous substrate-trapping mutant in PTP-PEST does not interact with the EGFR. This figure shows an anti-pTyr blot (RC20b) of precipitates prepared from lysates of COS cells that were transfected to express PTP1B, the C215A-PTP1B mutant (M1), the D181A-PTP1B mutant (M2), the GST fusions of these proteins, or PTP-PEST and the analogous mutants of it: C231S-PTP-PEST (M1) or D199A-PTP-PEST (M2). PTP-PEST was immunoprecipitated with mAb AG25 (27). The expression levels of the PTP-PEST proteins were similar to those for PTP1B.
Similar articles
- Design and characterization of an improved protein tyrosine phosphatase substrate-trapping mutant.
Xie L, Zhang YL, Zhang ZY. Xie L, et al. Biochemistry. 2002 Mar 26;41(12):4032-9. doi: 10.1021/bi015904r. Biochemistry. 2002. PMID: 11900546 - Epidermal growth factor receptor and the adaptor protein p52Shc are specific substrates of T-cell protein tyrosine phosphatase.
Tiganis T, Bennett AM, Ravichandran KS, Tonks NK. Tiganis T, et al. Mol Cell Biol. 1998 Mar;18(3):1622-34. doi: 10.1128/MCB.18.3.1622. Mol Cell Biol. 1998. PMID: 9488479 Free PMC article. - Protein tyrosine phosphatase 1B interacts with and is tyrosine phosphorylated by the epidermal growth factor receptor.
Liu F, Chernoff J. Liu F, et al. Biochem J. 1997 Oct 1;327 ( Pt 1)(Pt 1):139-45. doi: 10.1042/bj3270139. Biochem J. 1997. PMID: 9355745 Free PMC article. - Protein tyrosine phosphatase function: the substrate perspective.
Tiganis T, Bennett AM. Tiganis T, et al. Biochem J. 2007 Feb 15;402(1):1-15. doi: 10.1042/BJ20061548. Biochem J. 2007. PMID: 17238862 Free PMC article. Review. - The Extended Family of Protein Tyrosine Phosphatases.
Alonso A, Nunes-Xavier CE, Bayón Y, Pulido R. Alonso A, et al. Methods Mol Biol. 2016;1447:1-23. doi: 10.1007/978-1-4939-3746-2_1. Methods Mol Biol. 2016. PMID: 27514797 Review.
Cited by
- Central regulation of metabolism by protein tyrosine phosphatases.
Tsou RC, Bence KK. Tsou RC, et al. Front Neurosci. 2013 Jan 7;6:192. doi: 10.3389/fnins.2012.00192. eCollection 2012. Front Neurosci. 2013. PMID: 23308070 Free PMC article. - PECAM-1 is involved in BCR/ABL signaling and may downregulate imatinib-induced apoptosis of Philadelphia chromosome-positive leukemia cells.
Wu N, Kurosu T, Oshikawa G, Nagao T, Miura O. Wu N, et al. Int J Oncol. 2013 Feb;42(2):419-28. doi: 10.3892/ijo.2012.1729. Epub 2012 Dec 6. Int J Oncol. 2013. PMID: 23233201 Free PMC article. - Effect of oxidative stress on protein tyrosine phosphatase 1B in scleroderma dermal fibroblasts.
Tsou PS, Talia NN, Pinney AJ, Kendzicky A, Piera-Velazquez S, Jimenez SA, Seibold JR, Phillips K, Koch AE. Tsou PS, et al. Arthritis Rheum. 2012 Jun;64(6):1978-89. doi: 10.1002/art.34336. Epub 2011 Dec 12. Arthritis Rheum. 2012. PMID: 22161819 Free PMC article. - Regulation of myotubularin-related (MTMR)2 phosphatidylinositol phosphatase by MTMR5, a catalytically inactive phosphatase.
Kim SA, Vacratsis PO, Firestein R, Cleary ML, Dixon JE. Kim SA, et al. Proc Natl Acad Sci U S A. 2003 Apr 15;100(8):4492-7. doi: 10.1073/pnas.0431052100. Epub 2003 Mar 31. Proc Natl Acad Sci U S A. 2003. PMID: 12668758 Free PMC article. - A novel strategy for the development of selective active-site inhibitors of the protein tyrosine phosphatase-like proteins islet-cell antigen 512 (IA-2) and phogrin (IA-2beta).
Drake PG, Peters GH, Andersen HS, Hendriks W, Møller NP. Drake PG, et al. Biochem J. 2003 Jul 15;373(Pt 2):393-401. doi: 10.1042/BJ20021851. Biochem J. 2003. PMID: 12697028 Free PMC article.
References
- Tonks N K, Neel B G. Cell. 1996;87:365–368. - PubMed
- Tonks N K. Adv Pharmacol. 1996;36:91–119. - PubMed
- Barford D, Jia Z, Tonks N K. Nat Struct Biol. 1995;2:1043–1053. - PubMed
- Tonks N K, Diltz C D, Fischer E H. J Biol Chem. 1988;263:6722–6730. - PubMed
- Tonks N K, Diltz C D, Fischer E H. J Biol Chem. 1988;263:6731–6737. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous