The two alpha subunits of Escherichia coli RNA polymerase are asymmetrically arranged and contact different halves of the DNA upstream element - PubMed (original) (raw)
The two alpha subunits of Escherichia coli RNA polymerase are asymmetrically arranged and contact different halves of the DNA upstream element
K Murakami et al. Proc Natl Acad Sci U S A. 1997.
Abstract
RNA polymerase core enzyme of Escherichia coli is composed of two alpha subunits and one each of the beta and beta' subunits. The C-terminal domain of the RNA polymerase alpha subunit plays a key role in molecular communications with class I transcription factors and upstream (UP) elements of promoter DNA, using the same protein surface. To identify possible differences in the functional roles of the two alpha subunits, we have developed a reconstitution method for hybrid RNA polymerases containing two distinct alpha subunit derivatives in a defined orientation ("oriented alpha-heterodimer"). The binding sites of two alpha C-terminal domains on the UP element DNA were determined by hydroxyl radical-based DNA cleavage mediated by (p-bromoacetamidobenzyl)-EDTA x Fe, which was bound at Cys-269 on the UP recognition surface of one or both alpha subunits. The results clearly indicated that the two alpha subunits bind in tandem to two helix turns of the rrnBP1 UP element, and that the beta'-associated alpha subunit is bound to the promoter-distal region.
Figures
Figure 1
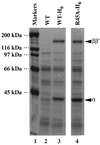
Assembly of mutant [45A]α into RNA polymerase. Extracts of E. coli cells expressing wild-type α (lane 2), His6-tagged α (lane 3), or His6-tagged mutant [45A]α (lane 4) were fractionated by chromatography on Ni2+-chelating columns. Bound proteins were eluted with a buffer containing 300 mM imidazole and fractionated by SDS/9% PAGE. The gel was stained with Coomassie brilliant blue.
Figure 2
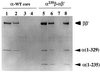
Subunit composition of the reconstituted RNA polymerases. A hybrid RNA polymerase with the subunit composition α235β-αβ′ was reconstituted from isolated β, β′, [45A]α with the His6 tag, and α-235 subunits. Reconstituted wild-type core enzyme (lanes 1–4) and the hybrid enzyme (lanes 5–8) were subjected to Ni2+ column chromatography as described. An aliquot of each fraction was analyzed by SDS/10% PAGE and Coomassie brilliant blue staining. Lanes: 1 and 5, the samples loaded; 2 and 6, flow-through fractions; 3 and 7, wash fractions; 4 and 8, fractions eluted with imidazole.
Figure 3
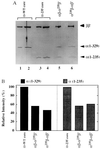
Stoichiometry of the two α subunits in the reconstituted RNA polymerases. (A) Reconstituted and purified RNA polymerase core enzymes with various combinations of α subunits were analyzed by electrophoresis on an SDS/10% polyacrylamide gel and stained with Coomassie brilliant blue. Lanes: 1 and 2, wild-type core enzyme isolated from E. coli cells; 3 and 4, the reconstituted core enzyme carrying α-235 homodimers; 5 and 6, the hybrid core enzymes containing α heterodimers in the defined orientations indicated above the lanes. The amounts of core enzyme analyzed were: lanes 1, 3, 5, and 6, 1 μg; lanes 2 and 4, 2 μg. (B) Band intensities of the full-length α [α(1–329)] or α-235 [α(1–235)] subunits were quantified and normalized against those of ββ′ subunit bands. The value of 100% represents 2 mol of α subunits per mol each of β and β′ subunits.
Figure 4
Western blotting of the reconstituted RNA polymerases cross-linked with DMS. The reconstituted RNA polymerase holoenzymes were treated with DMS and fractionated by SDS/gel electrophoresis. Gels were treated for immunostaining using anti-α (lanes 1–4), anti-HA (lanes 5–8), and anti-β (lanes 9–11) antibodies. Lanes 1, 2, 5, 6, and 9, holoenzyme with α-WT; lanes 3, 7, and 10, hybrid holoenzyme having β′ subunit-associated HA-tagged α (αβ-[45A]αHAβ′); lanes 4, 8, and 11, hybrid holoenzymes with β subunit-associated HA-tagged α (αHAβ-[45A]αβ′). Lanes 1 and 5 include the control unmodified samples. Subunit monomers (α and β) and cross-linked subunit complexes (α-α and α-β) are shown on the left, whereas the migration positions of molecular weight marker proteins are shown on the right.
Figure 5
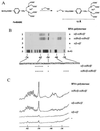
DNA cleavage by RNA polymerase-bound Fe·BABE. (A) Structure of Fe·BABE and its conjugation to a cysteine residue in RNA polymerase α subunit. The length of the reagent is about 12 Å. (B) DNA cleavage by the RNA polymerase having Fe·BABE at Cys-269 of the α subunit. A DNA fragment, 32P-end labeled on the top strand of _rrnB_P1, was incubated with RNA polymerase reconstituted from Fe·BABE-conjugated α subunits and then subjected to DNA cleavage by adding sodium ascorbate. Lanes: 1, DNA cleavage by the hybrid RNA polymerase modified by Fe·BABE only on the β′-associated α subunits [αβ-α(Fe)β′]; 2, DNA cleavage by the RNA polymerase having both α subunit Fe·BABE modified [α(Fe)β-α(Fe)β′]; 3, control reaction using wild-type RNA polymerase; 4, control reaction without RNA polymerase; 5, A+G-specific Maxam–Gilbert sequence. The nucleotide sequence shown at the bottom represents the top strand of _rrnB_P1. Asterisks show the main cleavage sites. (C) Cleavage patterns in B were scanned with a Bioimage Analyzer BAS2000 (Fujix).
Similar articles
- Mode of DNA-protein interaction between the C-terminal domain of Escherichia coli RNA polymerase alpha subunit and T7D promoter UP element.
Ozoline ON, Fujita N, Ishihama A. Ozoline ON, et al. Nucleic Acids Res. 2001 Dec 15;29(24):4909-19. doi: 10.1093/nar/29.24.4909. Nucleic Acids Res. 2001. PMID: 11812819 Free PMC article. - Transcription factor recognition surface on the RNA polymerase alpha subunit is involved in contact with the DNA enhancer element.
Murakami K, Fujita N, Ishihama A. Murakami K, et al. EMBO J. 1996 Aug 15;15(16):4358-67. EMBO J. 1996. PMID: 8861963 Free PMC article. - Role of the RNA polymerase alpha subunit in transcription activation.
Ishihama A. Ishihama A. Mol Microbiol. 1992 Nov;6(22):3283-8. doi: 10.1111/j.1365-2958.1992.tb02196.x. Mol Microbiol. 1992. PMID: 1484484 Review. - Escherichia coli RNA polymerase holoenzyme: rapid reconstitution from recombinant alpha, beta, beta', and sigma subunits.
Tang H, Kim Y, Severinov K, Goldfarb A, Ebright RH. Tang H, et al. Methods Enzymol. 1996;273:130-4. doi: 10.1016/s0076-6879(96)73012-4. Methods Enzymol. 1996. PMID: 8791605 Review. No abstract available.
Cited by
- Role of the RNA polymerase alpha subunits in CII-dependent activation of the bacteriophage lambda pE promoter: identification of important residues and positioning of the alpha C-terminal domains.
Kedzierska B, Lee DJ, Wegrzyn G, Busby SJ, Thomas MS. Kedzierska B, et al. Nucleic Acids Res. 2004 Feb 3;32(2):834-41. doi: 10.1093/nar/gkh230. Print 2004. Nucleic Acids Res. 2004. PMID: 14762211 Free PMC article. - Francisella RNA polymerase contains a heterodimer of non-identical α subunits.
Mukhamedyarov D, Makarova KS, Severinov K, Kuznedelov K. Mukhamedyarov D, et al. BMC Mol Biol. 2011 Nov 22;12:50. doi: 10.1186/1471-2199-12-50. BMC Mol Biol. 2011. PMID: 22108176 Free PMC article. - Requirement for two copies of RNA polymerase alpha subunit C-terminal domain for synergistic transcription activation at complex bacterial promoters.
Lloyd GS, Niu W, Tebbutt J, Ebright RH, Busby SJ. Lloyd GS, et al. Genes Dev. 2002 Oct 1;16(19):2557-65. doi: 10.1101/gad.237502. Genes Dev. 2002. PMID: 12368266 Free PMC article. - Mode of DNA-protein interaction between the C-terminal domain of Escherichia coli RNA polymerase alpha subunit and T7D promoter UP element.
Ozoline ON, Fujita N, Ishihama A. Ozoline ON, et al. Nucleic Acids Res. 2001 Dec 15;29(24):4909-19. doi: 10.1093/nar/29.24.4909. Nucleic Acids Res. 2001. PMID: 11812819 Free PMC article.
References
- Ishihama A. Adv Biophys. 1981;14:1–5. - PubMed
- Igarashi K, Fujita N, Ishihama A. J Mol Biol. 1991;218:1–6. - PubMed
- Kimura M, Fujita N, Ishihama A. J Mol Biol. 1994;242:107–115. - PubMed
- Kimura M, Ishihama A. J Mol Biol. 1995;248:756–767. - PubMed
- Kimura M, Ishihama A. J Mol Biol. 1995;254:342–349. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases