A hierarchy of Hu RNA binding proteins in developing and adult neurons - PubMed (original) (raw)
A hierarchy of Hu RNA binding proteins in developing and adult neurons
H J Okano et al. J Neurosci. 1997.
Abstract
The Hu proteins are a group of antigens targeted in an immune-mediated neurodegenerative disorder associated with cancer. We have cloned and characterized four members of the Hu gene family from mouse. We find that the Hu genes encode a large number of alternatively spliced transcripts to produce a series of related neuron-specific RNA binding proteins. Despite this complexity, we have discerned several ordered features of Hu expression. In the embryo, specific Hu genes are expressed in a hierarchy during early neurogenesis. In the E16 developing cortex, mHuB is induced in very early postmitotic neurons exiting the ventricular zone, mHuD is expressed in migrating neurons of the intermediate zone, and mHuC is expressed in mature cortical plate neurons. Such a hierarchy suggests distinct functional roles for each gene in developing neurons. In the adult, all neurons express some set of Hu mRNA and protein. However, specific patterns are evident such that individual neuronal types in the hippocampus, cerebellum, olfactory cortex, neocortex, and elsewhere express from one to several Hu genes. The complexity of potential protein variants within a gene family and of different Hu family members within a neuron suggests a diverse array of function. Given the strong homologies among the Hu proteins, the Drosophila neurogenic gene elav, and the Drosophila splicing factor sxl, we predict that different combinations of Hu proteins determine different neuron-specific aspects of post-transcriptional RNA regulation. Our findings of specific developmental patterns of expression and the correlation between immune targeting of the Hu proteins and adult neurodegenerative disease suggest that the Hu proteins are critical in both the proper development and function of mature neurons.
Figures
Fig. 1.
A, Amino acid alignments of Hu-related proteins. The deduced amino acid sequence for four mouse_Hu_ genes (mHuA, mHuB, mHuC, mHuD) is compared with the Drosophila elav and sxl_genes. Residues identical to the consensus are shown in bold type, and conservative substitutions are shown_gray-shaded. The extent of RNA recognition motifs (RRM) 1–3 is indicated.B, Sequence distances between mHu proteins and the_Drosophila elav_ and sxl proteins.Numbers shown represent percentage of similarity or percentage of divergence between the sequences shown in_A_. Amino acid sequences were aligned by using the Megalign program in the DNASTAR software package. C, Phylogenetic tree comparing the ancestral relationships between the mHu proteins and the Drosophila elav and sxl_proteins. The scale beneath the tree measures sequence distances. The sequence relationships were determined by using the Megalign program to compare the sequences shown in_A.
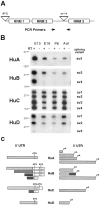
Fig. 2.
Alternative splicing of mouse Hu_transcripts. A, Location of alternative splicing in_Hu transcripts and the origin of primers for PCR. Two alternative splicing sites are shown. The site labeled_ATG_ includes splice variants encoding alternative start codons, and the site labeled sv1–4 includes splice variants within the spacer region between RRM 2 and_RRM 3_. B, Developmental analysis of splice variants sv1–4 by RT-PCR. Total RNA from the mouse brain of indicated developmental stage was reverse-transcribed with (lanes marked +) or without (lanes marked −) RT and amplified by PCR with gene-specific primers, the locations of which are indicated by_arrows_ in A. The name of each splicing variant is indicated and corresponds with the labels used in Table 1. Because sv2A and sv2B of HuB are not able to be distinguished by RT-PCR, both simply are named as _sv2_here. DNA size markers are shown on the left (in bp).C, Schematic drawing of Hu UTR alternative splice variants. ATG indicates putative initiation codons, and pA indicates polyadenylation sites found in cDNA clones. There is no apparent correlation between 5′ or 3′ UTR sequences between Hu families.
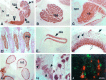
Fig. 3.
Distribution of Hu immunoreactivity in the E14 and adult mouse. A, Sagittal section (11 μm) from E14 mouse stained with affinity-purified Hu antiserum. There is intense immunoreactivity in the central and peripheral nervous systems, including the telencephalon, cerebellum, and spinal cord, but no reactivity in other tissues, including liver and heart.B, Hu immunoreactivity in E14 trigeminal ganglia.C, Hu immunoreactivity in E14 cerebellum, demonstrating intense staining in the developing cerebellum with no staining of the choroid plexus. D, Hu immunoreactivity in E14 dorsal root ganglia. E, Hu immunoreactivity in E14 olfactory epithelium. F, Hu immunoreactivity in E14 retina; Hu staining is intense in ganglion cell layer (open arrow) but absent in the ventricular surface (solid arrow), except in some scattered cells (arrowheads).G, Hu expression in the ganglion cells in the small intestine. H, High-power magnification of Hu immunoreactivity in E14 cortex. Note the cytoplasmic staining in the developing cells (arrowheads). I, High-power magnification of Hu immunoreactivity in a horizontal section (11 μm) of adult cortex, demonstrating that Hu reactivity in the differentiated neuron is both nuclear and cytoplasmic (arrows). J, Immunofluorescence double exposure of GFAP (green) and Hu(red) immunoreactivity in a horizontal section of adult cortex, demonstrating that the two are mutually exclusive.tl, Telencephalon; tgg, trigeminal ganglia; sc, spinal cord; cb, cerebellum;ht, heart; lv, liver; cpx, choroid plexus; drg, dorsal root ganglia;ofe, olfactory epithelium; int, small intestine. Scale bars: 2 mm in A; 160 μm in_B–E_, G; 80 μm in F; 20 μm in H–J.
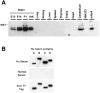
Fig. 4.
Western blot analysis of Hu antigen expression_in vivo_ and in vitro. A, Hu expression in the developing mouse brain and various adult tissues. Total cellular extracts from the indicated tissues, National Institutes of Health 3T3 cells, or purified mouse HuA fusion protein were run on 10% SDS-PAGE, transferred to nitrocellulose, and probed with Hu antiserum. The blot also was probed with an anti-tubulin antibody, which revealed that each of the nonbrain samples had at least as much (in most cases greater) protein loaded as in the lanes marked Brain or_Cerebellum_ (data not shown). B, Hu antiserum recognizes recombinant fusion proteins of all four Hu family members. Equal amounts of T7-tagged bacterial fusion proteins of mouse_HuA_, HuB, HuC, and_HuD_ were probed with paraneoplastic Hu antiserum (Hu Serum). The blot also was probed with normal human serum (Normal Serum), which was not reactive with the Hu fusion proteins, and an anti-T7 tag monoclonal antibody (Anti T7 Tag) used as a positive control and used to normalize the quantity of fusion protein present in each sample. Identical results were obtained with three different Hu disease antisera and Hu antisera affinity-purified with HuC fusion protein (data not shown).
Fig. 5.
Northern blot analysis of_HuA_ and HuC in various mouse tissues. Fifteen micrograms of total RNA from the indicated adult mouse tissues were separated on an agarose-formaldehyde gel, transferred onto nylon membranes, and hybridized with 32P-labeled 3′ UTR cDNA probes specific for mHuA or_mHuC_. RNAs were visualized with ethidium bromide for examining the equivalent amount of each sample (data not shown).
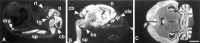
Fig. 6.
Specific expression of mHuC in the nervous system; dark-field microscopy of _HuC in situ_hybridization in the mouse. Sagittal sections (11 μm) of E16 (A) and P0 (B) mouse and a horizontal section (11 μm) of adult mouse brain (C) were hybridized with 33P-labeled antisense_mHuC_-specific cRNA probe. Mouse _HuC_expression is observed in the telencephalon, cerebellum, spinal cord, dorsal root ganglia, and olfactory epithelium at E16 and P0 and is absent in non-neural tissues. Expression in the nervous system persists through adulthood (C) and remains absent in non-neural tissue (data not shown). No reactivity was observed with a sense riboprobe (data not shown). bs, Brain stem;tg, trigeminal ganglia; rt, retina;tl, telencephalon; cb, cerebellum;drg, dorsal root ganglia; ofe, olfactory epithelium; sp, spinal cord. Scale bar, 2 mm.
Fig. 7.
Expression patterns of Hu mRNAs in developing brain. Sagittal sections (11 μm) of E14 mouse cortex (Ctx; A–D) and P9 mouse cerebellum (Cb; E–H) were analyzed by immunohistochemistry (A, E) and in situ_hybridization (B–D, F–H). Affinity-purified Hu antibody was used for immunohistochemistry (A, E). Serial sections were hybridized with 33P-labeled antisense_HuB (B, F), HuC(C, G), and HuD (D, H) gene-specific 3′ UTR cRNA probes. In the developing cortex, mHuB is expressed in some cells of the ventricular zone and cells of the intermediate zone;mHuB diminishes in the cortical plate, with no expression evident in the outermost differentiated neurons (arrowheads). mHuC is detected only in the cortical plate, including the differentiated neurons (arrowheads). mHuD expression is intense in the intermediate zone, diminishes in the cortical plate, and is very weak or absent in the differentiated neurons (arrowheads). In developing cerebellum,mHuB is expressed primarily in the external granule cell layer, whereas the expression of mHuC and_mHuD_ is distributed widely. Purkinje cells (arrows) express only mHuC. Sections were counterstained with cresyl violet. ml, Marginal layer;cp, cortical plate; iz, intermediate zone; vz, ventricular zone; egl, external germinal cell layer; m, molecular layer;p, Purkinje cell layer; igl, internal granule cell layer. Scale bars: 60 μm in A–D; 15 μm in E–H.
Fig. 8.
In situ hybridization of_Hu_ mRNAs in adult mouse brain and spinal cord. Horizontal sections (11 μm) of adult mouse brain and spinal cord were hybridized with 33P-labeled antisense probes specific for_mHuB_ (A, D, G, J, M, P),mHuC (B, E, H, K, N, Q), and_mHuD_ (C, F, I, L, O, R) cRNA probes. Hippocampal formation (Hf), olfactory bulb (Ob), cerebral cortex (Ctx), habenula (Hb), spinal cord (Sc), and cerebellum (Cb) are shown. Note that the hybridization signal of mHuB is intense in hippocampal pyramidal cells CA2, CA3, and CA4, mitral cell layer (mt), accessory olfactory bulb (ao), and dorsal root ganglia, but the signal is absent in the cerebellum except for some scattered cells in the granule cell layer (gr). The mRNA of mHuC is widely expressed in the adult nervous system, including hippocampal dentate gyrus (dg), pyramidal cells, glomerular (gl), mitral and granule cell layer (gr) of olfactory bulb, cortex, corpus striatum (st), medial habenula (mh), gray (gm) and white matter (wm) of spinal cord, dorsal root ganglia (drg), and Purkinje cells (p). mHuD expression is prominent in the entorhinal cortex (er), medial habenula, and dorsal root ganglia, but it is absent in dentate gyrus, corpus striatum, and Purkinje cells. No reactivity was observed with sense riboprobes (data not shown). v, Third ventricle;mol, molecular layer. Scale bars: 400 μm in_A_–C; 300 μm in_D–F_; 200 μm in G–I; 300 μm in_J–L;_ 400 μm in M–O; 100 μm in_P–R_.
Similar articles
- Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems.
Akamatsu W, Okano HJ, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell RB, Okano H. Akamatsu W, et al. Proc Natl Acad Sci U S A. 1999 Aug 17;96(17):9885-90. doi: 10.1073/pnas.96.17.9885. Proc Natl Acad Sci U S A. 1999. PMID: 10449789 Free PMC article. - Regulation of neuronal RNA signatures by ELAV/Hu proteins.
Hilgers V. Hilgers V. Wiley Interdiscip Rev RNA. 2023 Mar;14(2):e1733. doi: 10.1002/wrna.1733. Epub 2022 Apr 15. Wiley Interdiscip Rev RNA. 2023. PMID: 35429136 Review. - Transient expression of the conserved zinc finger gene INSM1 in progenitors and nascent neurons throughout embryonic and adult neurogenesis.
Duggan A, Madathany T, de Castro SC, Gerrelli D, Guddati K, García-Añoveros J. Duggan A, et al. J Comp Neurol. 2008 Apr 1;507(4):1497-520. doi: 10.1002/cne.21629. J Comp Neurol. 2008. PMID: 18205207 - ELAV proteins along evolution: back to the nucleus?
Colombrita C, Silani V, Ratti A. Colombrita C, et al. Mol Cell Neurosci. 2013 Sep;56:447-55. doi: 10.1016/j.mcn.2013.02.003. Epub 2013 Feb 22. Mol Cell Neurosci. 2013. PMID: 23439364 Review.
Cited by
- All three RNA recognition motifs and the hinge region of HuC play distinct roles in the regulation of alternative splicing.
Hinman MN, Zhou HL, Sharma A, Lou H. Hinman MN, et al. Nucleic Acids Res. 2013 May;41(9):5049-61. doi: 10.1093/nar/gkt166. Epub 2013 Mar 21. Nucleic Acids Res. 2013. PMID: 23525460 Free PMC article. - RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing.
Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, Dauksaite V, Vakeel P, Klaassen S, Gerull B, Thierfelder L, Regitz-Zagrosek V, Hacker TA, Saupe KW, Dec GW, Ellinor PT, MacRae CA, Spallek B, Fischer R, Perrot A, Özcelik C, Saar K, Hubner N, Gotthardt M. Guo W, et al. Nat Med. 2012 May;18(5):766-73. doi: 10.1038/nm.2693. Nat Med. 2012. PMID: 22466703 Free PMC article. - hiPSC-Based Model of Prenatal Exposure to Cannabinoids: Effect on Neuronal Differentiation.
Miranda CC, Barata T, Vaz SH, Ferreira C, Quintas A, Bekman EP. Miranda CC, et al. Front Mol Neurosci. 2020 Jul 6;13:119. doi: 10.3389/fnmol.2020.00119. eCollection 2020. Front Mol Neurosci. 2020. PMID: 32733202 Free PMC article. - Expression and distribution of HuR during ATP depletion and recovery in proximal tubule cells.
Jeyaraj SC, Dakhlallah D, Hill SR, Lee BS. Jeyaraj SC, et al. Am J Physiol Renal Physiol. 2006 Dec;291(6):F1255-63. doi: 10.1152/ajprenal.00440.2005. Epub 2006 Jun 20. Am J Physiol Renal Physiol. 2006. PMID: 16788138 Free PMC article. - Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock.
Gallouzi IE, Brennan CM, Steitz JA. Gallouzi IE, et al. RNA. 2001 Sep;7(9):1348-61. doi: 10.1017/s1355838201016089. RNA. 2001. PMID: 11565755 Free PMC article.
References
- Abitbol M, Menini C, Delezoide A, Rhyner T, Vekemans M, Mallet J. Nucleus basalis magnocellularis and hippocampus are the major sites of FMR-1 expression in the human fetal brain. Nat Genet. 1993;4:147–153. - PubMed
- Barami K, Iversen K, Furneaux H, Goldman SA. Hu protein as an early marker of neuronal phenotypic differentiation by subependymal zone cells of the adult songbird forebrain. J Neurobiol. 1995;28:82–101. - PubMed
- Bellen HJ, Kooyer S, D’Evelyn D, Pearlman J. The Drosophila couch potato protein is expressed in nuclei of peripheral neuronal precursors and shows homology to RNA-binding proteins. Genes Dev. 1992;6:2125–2136. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases