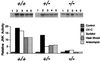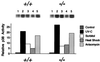Targeted disruption of the MKK4 gene causes embryonic death, inhibition of c-Jun NH2-terminal kinase activation, and defects in AP-1 transcriptional activity - PubMed (original) (raw)
Targeted disruption of the MKK4 gene causes embryonic death, inhibition of c-Jun NH2-terminal kinase activation, and defects in AP-1 transcriptional activity
D Yang et al. Proc Natl Acad Sci U S A. 1997.
Abstract
MKK4 is a member of the mitogen-activated protein kinase kinase group of dual specificity protein kinases that functions as an activator of the c-Jun NH2-terminal kinase (JNK) in vitro. To examine the function of MKK4 in vivo, we investigated the effect of targeted disruption of the MKK4 gene. Crosses of heterozygous MKK4 (+/-) mice demonstrated that homozygous knockout (-/-) animals die before embryonic day 14, indicating that the MKK4 gene is required for viability. The role of MKK4 in JNK activation was examined by investigation of cultured MKK4 (+/+) and MKK4 (-/-) cells. Disruption of the MKK4 gene blocked JNK activation caused by: (i) the mitogen-activated protein kinase kinase kinase MEKK1, and (ii) treatment with anisomycin or heat shock. In contrast, JNK activation caused by other forms of environmental stress (UV-C radiation and osmotic shock) was partially inhibited in MKK4 (-/-) cells. Regulated AP-1 transcriptional activity, a target of the JNK signal transduction pathway, was also selectively blocked in MKK4 (-/-) cells. Complementation studies demonstrated that the defective AP-1 transcriptional activity was restored by transfection of MKK4 (-/-) cells with an MKK4 expression vector. These data establish that MKK4 is a JNK activator in vivo and demonstrate that MKK4 is an essential component of the JNK signal transduction pathway.
Figures
Figure 1
Strategy for the targeted disruption of the MKK4 gene. The pBSNTK2 vector, the MKK4 targeting vector, the genomic region at the MKK4 locus, and the predicted structure of the mutated MKK4 gene. Restriction enzyme sites are indicated (B, _Bam_HI; R, _Eco_RI; N, _Not_I; X, _Xho_I; Ap, _Apa_I; Sp, _Spe_I; Xb, _Xba_I). The two hatched boxes are MKK4 exons corresponding to the subdomain VII and VIII (amino acid residues 211–268). SIAKT is the sequence (single-letter code) that includes the dual phosphorylation sites that are required for MKK4 activation.
Figure 2
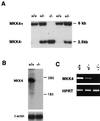
Identification of ES cell clones with targeted disruption of the MKK4 gene. (A) Genomic DNA prepared from ES cell clones indicates the presence of all three expected genotypes. _Eco_RI restricted DNA was probed with a radiolabeled fragment of the MKK4 gene (Fig. 1). The upper band (6 kb) corresponds to the wild-type allele, and the lower band (2.5 kb) corresponds to the mutant allele. (B) Northern blot analysis of total RNA (10 μg) isolated from wild-type and homozygous knockout ES cells. Blots were probed with a random-primed 32P-labeled mouse MKK4 cDNA probe. The blots also were probed for β-actin mRNA as an internal control. (C) Reverse transcription-PCR analysis of MKK4 mRNA expressed by ES cells. RNA isolated from ES cells was amplifed with primers specific for MKK4 and hypoxanthine–guanine phosphoribosyltransferase mRNA.
Figure 3
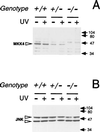
Expression of MKK4 and JNK in ES cell clones. Lysates (30 μg) prepared from wild-type (+/+), heterozygous (+/−), and homozygous knockout (−/−) MKK4 cells were examined by Western blot analysis using antibodies to MKK4 (A) and JNK (B). The cells were treated without (−) and with (+) UV-C radiation (80 J/m2) and harvested after incubation at 37°C for 1 h. Open arrows indicate MKK4 (45 kDa) and JNK (46 kDa and 55 kDa). Closed arrows indicate the migration of molecular mass standards (kDa).
Figure 4
Effect of MKK4 gene disruption on JNK activation. Wild-type (+/+), heterozygous (+/−), and homozygous knockout (−/−) MKK4 cells were untreated (lane 1) or treated with (lane 2) UV-C (80 J/m2), (lane 3) osmotic shock (300 mM sorbitol), (lane 4) heat shock (42°C, 15 min.), or (lane 5) anisomycin (5 μg/ml). The cells were harvested after incubation at 37°C for 1 h. JNK activity was measured using an immunecomplex kinase assay with the substrate GST-cJun. The radioactivity incorporated into GST-cJun was quantitated after SDS/PAGE by Phosphorimager analysis.
Figure 5
Effect of MKK4 gene disruption on p38 MAP kinase activation. Wild-type (+/+) and homozygous knockout (−/−) MKK4 cells were untreated (lane 1) or treated with (lane 2) UV-C (80 J/m2), (lane 3) osmotic shock (300 mM sorbitol), (lane 4) heat shock (42°C, 15 min), or (lane 5) anisomycin (5 μg/ml). The cells were harvested after incubation at 37°C for 1 h. p38 MAP kinase activity was measured using an immunecomplex kinase assay with the substrate GST-ATF2. The radioactivity incorporated into GST-ATF2 was quantitated after SDS/PAGE by PhosphorImager analysis.
Figure 6
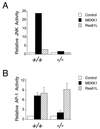
MKK4 is required for MEKK1-induced JNK activation and AP-1 transcriptional activity. (A) Wild-type (+/+) and homozygous knockout (−/−) MKK4 cells were cotransfected with pCDNA3-Flag-JNK1 together with pCMV5, pCMV5-MEKK1, or pCMV-Ras61L. The epitope-tagged JNK1 was immunoprecipitated with the M2 monoclonal antibody. Protein kinase activity in the immunecomplexes was measured using the substrate GST-cJun. The radioactivity incorporated into GST-cJun was quantitated after SDS/PAGE by Phosphorimager analysis. (B) Wild-type (+/+) and homozygous knockout (−/−) MKK4 cells were cotransfected with the TRE-luciferase reporter plasmid together with activated Ras or MEKK1. Transfection efficiency was monitored using the β-galactosidase expression vector pCH110. The relative AP-1-dependent reporter gene expression is presented as the activity ratio of luciferase/β-galactosidase. Data are the mean ± SD (n = 3).
Figure 7
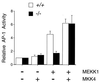
Complementation analysis of MKK4 gene disruption. Wild-type (+/+) and homozygous knockout (−/−) MKK4 cells were cotransfected with the TRE-luciferase reporter plasmid together with MKK4 or MEKK1 expression vectors, as indicated. Transfection efficiency was monitored using the β-galactosidase expression vector pCH110. The relative AP-1-dependent reporter gene expression is presented as the activity ratio of luciferase/β-galactosidase. Data are the mean ± SD (n = 3).
Similar articles
- Coordinate activation of activator protein 1 and inflammatory cytokines in response to Neisseria gonorrhoeae epithelial cell contact involves stress response kinases.
Naumann M, Rudel T, Wieland B, Bartsch C, Meyer TF. Naumann M, et al. J Exp Med. 1998 Oct 5;188(7):1277-86. doi: 10.1084/jem.188.7.1277. J Exp Med. 1998. PMID: 9763607 Free PMC article. - A novel SAPK/JNK kinase, MKK7, stimulated by TNFalpha and cellular stresses.
Moriguchi T, Toyoshima F, Masuyama N, Hanafusa H, Gotoh Y, Nishida E. Moriguchi T, et al. EMBO J. 1997 Dec 1;16(23):7045-53. doi: 10.1093/emboj/16.23.7045. EMBO J. 1997. PMID: 9384583 Free PMC article. - Human HPK1, a novel human hematopoietic progenitor kinase that activates the JNK/SAPK kinase cascade.
Hu MC, Qiu WR, Wang X, Meyer CF, Tan TH. Hu MC, et al. Genes Dev. 1996 Sep 15;10(18):2251-64. doi: 10.1101/gad.10.18.2251. Genes Dev. 1996. PMID: 8824585 - Signal transduction by the c-Jun N-terminal kinase.
Davis RJ. Davis RJ. Biochem Soc Symp. 1999;64:1-12. doi: 10.1515/9781400865048.1. Biochem Soc Symp. 1999. PMID: 10207617 Review. - Mitogen-activated protein kinase kinase 4 (MKK4).
Cuenda A. Cuenda A. Int J Biochem Cell Biol. 2000 Jun;32(6):581-7. doi: 10.1016/s1357-2725(00)00003-0. Int J Biochem Cell Biol. 2000. PMID: 10785355 Review.
Cited by
- Epitranscriptomic reader YTHDF2 regulates SEK1(MAP2K4)-JNK-cJUN inflammatory signaling in astrocytes during neurotoxic stress.
Malovic E, Ealy A, Miller C, Jang A, Hsu PJ, Sarkar S, Rokad D, Goeser C, Hartman AK, Zhu A, Palanisamy B, Zenitsky G, Jin H, Anantharam V, Kanthasamy A, He C, Kanthasamy AG. Malovic E, et al. iScience. 2024 Jul 31;27(9):110619. doi: 10.1016/j.isci.2024.110619. eCollection 2024 Sep 20. iScience. 2024. PMID: 39252959 Free PMC article. - Mitogen-activated protein kinase kinase 7 in inflammatory, cancer, and neurological diseases.
Caliz AD, Vertii A, Fisch V, Yoon S, Yoo HJ, Keaney JF Jr, Kant S. Caliz AD, et al. Front Cell Dev Biol. 2022 Oct 20;10:979673. doi: 10.3389/fcell.2022.979673. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36340039 Free PMC article. Review. - Mitogen Kinase Kinase (MKK7) Controls Cytokine Production In Vitro and In Vivo in Mice.
Caliz AD, Yoo HJ, Vertii A, Dolan AC, Tournier C, Davis RJ, Keaney JF Jr, Kant S. Caliz AD, et al. Int J Mol Sci. 2021 Aug 29;22(17):9364. doi: 10.3390/ijms22179364. Int J Mol Sci. 2021. PMID: 34502275 Free PMC article. - Nonylphenol Induces Apoptosis through ROS/JNK Signaling in a Spermatogonia Cell Line.
Park HJ, Lee R, Yoo H, Hong K, Song H. Park HJ, et al. Int J Mol Sci. 2020 Dec 30;22(1):307. doi: 10.3390/ijms22010307. Int J Mol Sci. 2020. PMID: 33396729 Free PMC article. - The role of MKK4 in T-cell development and immunity to viral infections.
Preston SP, Doerflinger M, Scott HW, Allison CC, Horton M, Cooney J, Pellegrini M. Preston SP, et al. Immunol Cell Biol. 2021 Apr;99(4):428-435. doi: 10.1111/imcb.12426. Epub 2020 Dec 8. Immunol Cell Biol. 2021. PMID: 33175451 Free PMC article.
References
- Whitmarsh A J, Davis R J. J Mol Med. 1996;74:589–607. - PubMed
- Karin M. J Biol Chem. 1995;270:16483–16486. - PubMed
- Davis R J. Trends Biochem Sci. 1994;19:470–473. - PubMed
- Dérijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. Cell. 1994;76:1025–1037. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous