Restrictive clonal allocation in the chimeric mouse brain - PubMed (original) (raw)
Restrictive clonal allocation in the chimeric mouse brain
C Y Kuan et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Whether, and to what extent, lineage restriction contributes to the organization of the mammalian brain remains unclear. Here we address this issue by examining the distribution of clonally related cells in chimeric mice generated by injecting genetically tagged embryonic stem (ES) cells into blastocyst embryos. Our examination of postnatal chimeric brains revealed that the vast majority of labeled ES cell descendents were confined within a different subset of brain regions in each animal. Moreover, the deployment of labeled cells in different brain regions was distinctive. The pattern of ordered and binomial colonization suggested that early diversified founder cells may constrain the fates of their descendants through a restriction of dispersion. In addition, the symmetrical distribution of ES cell descendants suggests that bilaterally corresponding structures may arise from a common set of progenitor cells. Finally, clones of cells formed a continuous band within the deep strata of the neocortex. This later finding in conjunction with the radial distribution of clones in remaining layers observed in previous studies indicates that the cerebral neocortex may derive from two groups of founder cells, which is consistent with the hypothesis of dual phylogenetic origins of the mammalian cerebral cortex.
Figures
Figure 1
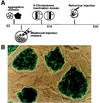
(A) Schematic illustration of the major approaches for retrospective clonal analysis in the mouse. These include: making aggregated embryos between genetically distinct strains at the 4- to 8-cell stage (12, 13); injecting genetically tagged ES cells into blastocyst embryos (14); taking advantage of random X chromosome inactivation of transgene in female mice (15); and infecting embryos with replication-defected retrovirus (–18). The timing of labeling clones in each method also is indicated in perspective. In the present study, D3 ES cells were first transfected and selected for stable expression of the lacZ gene and then used for injection into mouse blastocyst embryos. As shown in B, colonies of undifferentiated ES cells (actin 69) display homogeneous expression of the lacZ gene on top of a fibroblast feeder cell layer in vitro.
Figure 2
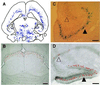
(A) The general distribution pattern is illustrated here by camera lucida plotting of labeled cells in chimera 4. Although complicated, the distribution pattern is not haphazard. Three distinguished features of distribution are illustrated in B, C, and D. First (B), labeled cells tend to occupy certain layers in laminated brain regions. For example, labeled cells (red—pseudocolored to highlight the position of cells at low magnification) are restricted to the outermost superficial gray layer in the superior colliculus in chimera 5. Second (C), the deployment of labeled cells in deferent brain regions is distinct. As shown here, labeled cells were distributed evenly along the outer margin of the dentate gyrus (filled triangle) but were scattered discontinuously in the nearby Ammon’s horn (unfilled triangle; chimera 4). Third (D), the same region-specific deployment of labeled cells can be found in other animals, such as the hippocampal region of chimera 1 shown here. (Bar = 100 μm.)
Figure 3
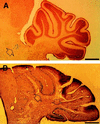
(A) Labeled cells were found in the brainstem (arrowhead) in chimera 1 but not in the cerebellum. (B) On the contrary, a densely packed population of ES cell descendants (arrowhead) was allocated in the cerebellum across several folia in chimera 11. Such a great magnitude of change of presence of labeled cells and their ordered alignment indicates a binomial colonization of ES cell descendants in a brain region. (Bar = 500 μm.)
Figure 4
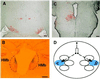
Bilaterally corresponding structures are typically both populated by ES cell descendants. (A) Although only a small number of labeled cells (pseudocolored to highlight the position of cells at low magnification) was found in this section, the cells were allocated symmetrically in the diencephalon (chimera 4). (B) The medial habenular nucleus (HMb) was not a preferentially incorporated structure, but, when labeled cells were located on one side, they were symmetrically localized on the opposite side of the habenular nucleus (chimera 9). (C) Nevertheless, a unilateral allocation of labeled cells was observed occasionally (chimera 8). (D) One possible mechanism to generate such bilaterally symmetric allocations is that cells of the same ancestry tend to incorporate into bilaterally corresponding structures. The sizes of bilaterally sibling clones, as indicated in the figure, are not necessarily expanded to the same extent. (Bar = 100 μm.)
Figure 5
(A) Camera lucida plotting of ES cell descendants from three sections of chimera 5 indicates a continuous laminar allocation restricted to the deep layers (V and VI) of the entire cerebral cortex. Labeled cells in the upper cortical plate were much scarcer and more sporadically distributed. (B) Double labeling with antibody against glial fibrillary acidic protein indicated that astrocytes (arrowhead) were only a small percentage of this group of _lacZ_+ cells located above the white matter (WM). (C) Tissue blocks of the deep and the upper cerebral cortices were picked individually from histological specimens and processed for PCR detection of the lacZ gene (200-bp signal). The endogenous γ-enteric actin gene (SMGA, 100-bp signal) also was amplified to ascertain the successful harvest of DNA from histologic specimens. Results of one representative set of samples are shown here. In a larger series of samples (n = 43), 72% of the deep cortical samples and 11.6% of the upper cortical samples were _lacZ_-positive. The correlation between X-Gal histochemical expression and the presence of the lacZ gene was statistically significant (P < 0.005 in χ2 analysis; df = 1). (D) Comparative studies indicated that the three-layered general cortex expanded and propelled the archicortex (A: hippocampus, H) and the paleocortex (P: piriform cortex, P) toward the medial wall of telencephalon during evolution. It also has been postulated that neuroblasts of the dorsal ventricular ridge (DVR) located in the dorsal striatum (S) migrated and incorporated into the general cortex in ancestral mammals to result in the modern six-layered isocortex (4). One possible explanation of the coexistence of radial and laminar clones in the mouse cerebral cortex is that these two phylogenetically distinct populations of cells remain ontogenetically segregated and are allocated differently. (D is modeled after ref. 4). (Bar = 100 μm.)
Comment in
- Evolutionary developmental biology meets the brain: the origins of mammalian cortex.
Karten HJ. Karten HJ. Proc Natl Acad Sci U S A. 1997 Apr 1;94(7):2800-4. doi: 10.1073/pnas.94.7.2800. Proc Natl Acad Sci U S A. 1997. PMID: 9096300 Free PMC article. No abstract available.
Similar articles
- Developmental fate of single embryonic stem cells microinjected into 8-cell-stage mouse embryos.
Saburi S, Azuma S, Sato E, Toyoda Y, Tachi C. Saburi S, et al. Differentiation. 1997 Oct;62(1):1-11. doi: 10.1046/j.1432-0436.1997.6210001.x. Differentiation. 1997. PMID: 9373942 - At most three ES cells contribute to the somatic lineages of chimeric mice and of mice produced by ES-tetraploid complementation.
Wang Z, Jaenisch R. Wang Z, et al. Dev Biol. 2004 Nov 1;275(1):192-201. doi: 10.1016/j.ydbio.2004.06.026. Dev Biol. 2004. PMID: 15464582 - Embryonic stem cells expressing both platelet endothelial cell adhesion molecule-1 and stage-specific embryonic antigen-1 differentiate predominantly into epiblast cells in a chimeric embryo.
Furusawa T, Ohkoshi K, Honda C, Takahashi S, Tokunaga T. Furusawa T, et al. Biol Reprod. 2004 May;70(5):1452-7. doi: 10.1095/biolreprod.103.024190. Epub 2004 Jan 21. Biol Reprod. 2004. PMID: 14736812 - Clonal analysis of early mammalian development.
Gardner RL. Gardner RL. Philos Trans R Soc Lond B Biol Sci. 1985 Dec 17;312(1153):163-78. doi: 10.1098/rstb.1985.0186. Philos Trans R Soc Lond B Biol Sci. 1985. PMID: 2869527 Review. - New approach for the establishment of mouse early embryonic stem cells and induction of their differentiation.
Ishiwata I, Tokeida Y, Iguchi M, Ishiwata C, Kiguchi K, Yasumoto S, Sato K, Tachibana T, Hashimoto H, Ishikawa H. Ishiwata I, et al. Hum Cell. 2001 Dec;14(4):283-91. Hum Cell. 2001. PMID: 11925930 Review.
Cited by
- Aggregation chimeras provide evidence of in vivo intercellular correction in ovine CLN6 neuronal ceroid lipofuscinosis (Batten disease).
Barry LA, Kay GW, Mitchell NL, Murray SJ, Jay NP, Palmer DN. Barry LA, et al. PLoS One. 2022 Apr 11;17(4):e0261544. doi: 10.1371/journal.pone.0261544. eCollection 2022. PLoS One. 2022. PMID: 35404973 Free PMC article. - Neurons lacking huntingtin differentially colonize brain and survive in chimeric mice.
Reiner A, Del Mar N, Meade CA, Yang H, Dragatsis I, Zeitlin S, Goldowitz D. Reiner A, et al. J Neurosci. 2001 Oct 1;21(19):7608-19. doi: 10.1523/JNEUROSCI.21-19-07608.2001. J Neurosci. 2001. PMID: 11567051 Free PMC article. - Cortical projection neurons: sprung from the same root.
Han W, Sestan N. Han W, et al. Neuron. 2013 Dec 4;80(5):1103-5. doi: 10.1016/j.neuron.2013.11.016. Neuron. 2013. PMID: 24314721 Free PMC article. - Clonal architecture of the mouse hippocampus.
Martin LA, Tan SS, Goldowitz D. Martin LA, et al. J Neurosci. 2002 May 1;22(9):3520-30. doi: 10.1523/JNEUROSCI.22-09-03520.2002. J Neurosci. 2002. PMID: 11978829 Free PMC article. - Evolutionary developmental biology meets the brain: the origins of mammalian cortex.
Karten HJ. Karten HJ. Proc Natl Acad Sci U S A. 1997 Apr 1;94(7):2800-4. doi: 10.1073/pnas.94.7.2800. Proc Natl Acad Sci U S A. 1997. PMID: 9096300 Free PMC article. No abstract available.
References
- Jacobson M. Developmental Neurobiology. 3rd Ed. New York: Plenum Press; 1991.
- Rakic P. Science. 1988;241:170–176. - PubMed
- Nauta W, Karten H. In: The Neurosciences: Second Study Program. Schmitt F, editor. New York: Rockefeller Univ. Press; 1970. pp. 7–26.
- Puelles L, Rubenstein J. Trends Neurosci. 1993;16:472–479. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources