cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin - PubMed (original) (raw)
cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin
F Chang et al. J Cell Biol. 1997.
Abstract
As in many other eukaryotic cells, cell division in fission yeast depends on the assembly of an actin ring that circumscribes the middle of the cell. Schizosaccharomyces pombe cdc12 is an essential gene necessary for actin ring assembly and septum formation. Here we show that cdc12p is a member of a family of proteins including Drosophila diaphanous, Saccharomyces cerevisiae BNI1, and S. pombe fus1, which are involved in cytokinesis or other actin-mediated processes. Using indirect immunofluorescence, we show that cdc12p is located in the cell division ring and not in other actin structures. When overexpressed, cdc12p is located at a medial spot in interphase that anticipates the future ring site. cdc12p localization is altered in actin ring mutants. cdc8 (tropomyosin homologue), cdc3 (profilin homologue), and cdc15 mutants exhibit no specific cdc12p staining during mitosis. cdc4 mutant cells exhibit a medial cortical cdc12p spot in place of a ring. mid1 mutant cells generally exhibit a cdc12p spot with a single cdc12p strand extending in a random direction. Based on these patterns, we present a model in which ring assembly originates from a single point on the cortex and in which a molecular pathway for the functions of cytokinesis proteins is suggested. Finally, we found that cdc12 and cdc3 mutants show a synthetic-lethal genetic interaction, and a proline-rich domain of cdc12p binds directly to profilin cdc3p in vitro, suggesting that one function of cdc12p in ring assembly is to bind profilin.
Figures
Figure 1
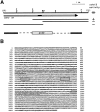
cdc12 encodes a protein similar to the dia/BNI1 family of proteins. (A) Localization of the cdc12 gene. Restriction map of the original 6-kb genomic clone insert (pFC103) that rescues cdc12-112 mutant cells. (Arrow) Extent and orientation of the single large open reading frame in the region. Truncated versions of the clone and their ability to complement a cdc12-112 mutant; ++, full rescue; +, partial rescue. cdc12::ura4 deletion construct used in deleting cdc12. Much of the cdc12 open reading frame is replaced with a 1.6-kb ura4 insert. (B) cdc12 open reading frame. Regions of predicted coiled-coil are underlined (Lupas et al., 1991), and the polyproline regions are boxed. (C) Similarity in gene domain structure. Proteins share a central FH1 polyproline rich domain (black box), an FH2 domain (stripe box), and a larger COOH-terminal area of homology (gray) also diagrammed in D. (D) Similarity of the proteins in a COOH-terminal region. Proteins were aligned using Megalign in DNA Star. Residues identical to a calculated consensus are colored in black, and conserved residues are colored in gray. These sequence data are available from GenBank/EMBL/DDBJ under accession number Q10059.
Figure 1
cdc12 encodes a protein similar to the dia/BNI1 family of proteins. (A) Localization of the cdc12 gene. Restriction map of the original 6-kb genomic clone insert (pFC103) that rescues cdc12-112 mutant cells. (Arrow) Extent and orientation of the single large open reading frame in the region. Truncated versions of the clone and their ability to complement a cdc12-112 mutant; ++, full rescue; +, partial rescue. cdc12::ura4 deletion construct used in deleting cdc12. Much of the cdc12 open reading frame is replaced with a 1.6-kb ura4 insert. (B) cdc12 open reading frame. Regions of predicted coiled-coil are underlined (Lupas et al., 1991), and the polyproline regions are boxed. (C) Similarity in gene domain structure. Proteins share a central FH1 polyproline rich domain (black box), an FH2 domain (stripe box), and a larger COOH-terminal area of homology (gray) also diagrammed in D. (D) Similarity of the proteins in a COOH-terminal region. Proteins were aligned using Megalign in DNA Star. Residues identical to a calculated consensus are colored in black, and conserved residues are colored in gray. These sequence data are available from GenBank/EMBL/DDBJ under accession number Q10059.
Figure 2
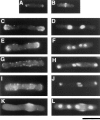
Phenotype of cdc12 + and cdc12::ura4 deletion cells. (A and B). Wild-type cells stained with rhodamine-phalloidin, showing normal interphase (A) and mitotic (B) actin patterns. (C–L) cdc12::ura4 cells stained with rhodamine-phallodin (C, E, G, and I), DAPI (D, F, H, and J), or calcofluor (K and L). Diploid ura4/ ura4 leu1/leu1 ade6-M16/ade6-M22 cdc12 +/cdc12::ura4 cells were sporulated, and the spores were germinated in media containing no uracil, so only cdc12::ura4 spores could germinate. Spores were grown for 15 h at 30°C, fixed, and stained. (C–F) cdc12::ura4 cells in interphase, showing normal interphase polarized actin distribution at the cell tips. Cell in D has two nuclei, and cell in F has proceeded through another generation and contains four nuclei. (G–J) cdc12::ura4 mitotic cells with actin dots instead of an actin ring. Cell in G is focused on the middle of the cell, while the cell in I is focused on the cell surface. Nuclear patterns in H and I are characteristic of cells in mitosis (see Fig. 6_A_ for comparison). Calcofluor-stained cells show no septum cell wall deposition even after multiple nuclear divisions. The rounded bulge that stains with Calcofluor (K and L) in cdc12::ura4 cells represents the site of the original spore wall. Bar, 10 μm.
Figure 6
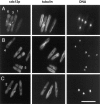
Localization of cdc12p in different actin ring mutants during mitosis. (A) cdc12-299 (FC127); (B) cdc3313 (FC114); (C) cdc8-346 (FC131); (D) cdc15-287 (FC55); (E and F) cdc4-377 (FC361). Mutant cells were grown in YE5S at permissive temperature (25°C), and then shifted to restrictive temperature (35.5°C) for 4 h. Cells were fixed and processed for immunofluorescence with anti-cdc12 antibody, anti-tubulin antibody or anti-actin antibody, and DAPI. On the basis of spindle and nuclear morphology, cells pictured are in anaphase, the cell cycle period of maximum cdc12p ring staining in wild-type cells. (A–C) Cells are in mitosis in which two nuclei are dividing into four and thus exhibit double spindles; (E–F) cells exhibit single spindles. (A–C) Mutants exhibit no specific cdc12p staining; the cytoplasmic staining is nonspecific background staining. (E and F) Note the medial spot of cdc12p (arrow) in the cdc4 mutant, which localizes to the origin of an actin aster (F, middle panel, labeled a). Bar, 10 μm.
Figure 3
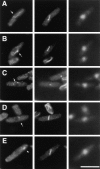
cdc12p localizes to the cell division ring in wild-type cells. (A) Specificity of the anti-cdc12 affinity-purified antibody. 5 μg of crude whole yeast cell extracts were analyzed by Western blotting using affinity-purified anti-cdc12 antibody. (Lane 1) Extract from wild-type cells. (Lane 2) Extract from cells overexpressing cdc12p. Cells transformed with the plasmid pnmt-cdc12 (FC291) were shifted to derepressing conditions (minimal media without thiamine) for 20 h at 30°C. Molecular weight markers (kD) are indicated. Serial dilutions showed that the pnmt-cdc12 extract contains 10–20-fold more cdc12p than the wild-type extracts (data not shown). (B–K) cdc12p localization in wild-type cells. Wild-type cells were grown in YE5S at 30°C in exponential phase, fixed, and processed for immunofluorescence using anti-cdc12 antibody and DAPI (see Materials and Methods for details). (B, D, F, H, and J) Anti-cdc12 staining. (C, E, G, I, and K) DAPI staining. (B and C) A typical field of cells containing cells from different phases of the cell cycle. Note examples of ring staining. (e) Cell in early mitosis in which the ring is beginning to form in an asymmetrical fashion. (i) Interphase with no specific cdc12 staining. (B, inset) Cross-section of a cdc12p ring. (D and E) Cell in early mitosis; (F and G) cell in early anaphase; (H and I) cell in midanaphase; (J and K) cell in postanaphase during ring closure. Patchy cytoplasmic staining is also seen in cells lacking cdc12p, and thus probably represents background staining (data not shown). Bar, 10 μm.
Figure 5
The positions of the cdc12p spot and the ring coincide. FC565 (pnmt-cdc12) cells were processed as described in Fig. 4. Shown are four representative cells in early mitosis, around the period of ring formation. Top three panels show three different focal planes of the same cell, and the bottom two panels show tubulin and DAPI staining as labeled. (A) Cell with a cdc12p spot. (B) Cell with a cdc12p associated with a complete ring. (C) Cell with a cdc12p spot with an incomplete ring. Note the discontinuity of the ring adjacent to the spot in the top panel. (D) Cell with a cdc12p ring but no spot. Bar, 10 μm.
Figure 4
Upon overexpression, cdc12p localizes to a medial spot. FC565 (pnmtcdc12) cells were induced in derepressing conditions for 20 h at 30°C, fixed in cold methanol, and processed for immunofluorescence with anti-cdc12 antibody (left panels), anti-tubulin antibody (middle panels), and DAPI (right panels). The tubulin and DAPI staining patterns identify the cell cycle stage. (A–C) All panels show three representative fields from the same population of cells. (Cell 1) Interphase cell with a medial cdc12p spot. (Cell 2) Early mitotic cell with a medial spot. (Cell 3) Cell in postanaphase with a cdc12p ring. (Cell 4) Cell in postanaphase with a closed ring. (Cell 5) Cell after cell division with a cdc12p spot at the presumed midbody on the cell tip. (Cell 6) Cell with two spots, one at the cell tip and one at the cell middle. Bar, 10 μm.
Figure 7
Localization of cdc12p in mid1 mutant cells during early mitosis. mid1-366 mutant cells were grown in YE5S at permissive temperature (25°C), and then shifted to restrictive temperature (35.5°C) for 2 h. Cells were processed for immunofluorescence with anti-cdc12 antibody, anti-tubulin antibody or anti-actin antibody, and DAPI. On the basis of spindle and nuclear morphology, cells pictured are in early mitosis, the cell cycle period around ring formation. Note different configurations of cdc12p dot and strand (arrows). Bar, 10 μm.
Figure 8
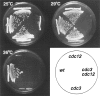
cdc3 and cdc12 exhibit a synthetic-lethal genetic interaction. Siblings from a tetratype tetrad in a cross between cdc3-313 and cdc12-112 mutants were streaked on YE5S plates and incubated at 25°, 29°, or 36°C. Growth was assayed by colony formation. Note that the cdc3 cdc12 double mutant exhibited markedly reduced growth at 25° and 29°C where the single mutants are viable. Strains were FC322 (cdc12-112), FC323 (wt), FC324 (cdc3313), and FC325 (cdc3-313 cdc12-112).
Figure 9
cdc12p fragment binds to profilin cdc3p in vitro. (A) Purified proteins used in these studies. (Lane 1) Purified GST-cdc3. (Lane 2) Purified GST. (Lane 3) Purified His-tagged cdc12p fragment. (B and C) cdc12p binds to GST-cdc3 but not to GST. In each 180-μl reaction, cdc12p (1.6 μM final concentration) and 20-μl 50% slurries of glutathione agarose beads containing indicated concentrations of GST-cdc3 or GST were added to each reaction. Reactions were incubated at room temperature for 1 h, and the beads were pelleted. (B) Coomassie-stained gel of total proteins before pelleting (lane 1) and pellet fractions (lanes 2–10) at equivalent volumes. Bands were quantitated by densitometry relative to total protein in lane 1 and plotted in C. (D and E) Poly-
l
-proline inhibits binding. Varying amounts of poly-
l
-proline were added to the binding reactions similar to ones described above except cdc12p was 0.8 μM. (D) Coomassie-stained gel of the pellet fractions. (E) cdc12p in the pellets was quantitated by densitometry relative to supernatant fractions (not shown).
Figure 10
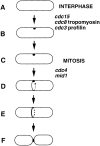
A model for the establishment of the division site in S. pombe. (A and B) In interphase, a marker spot is established at the cell middle, possibly by a signal from the nucleus. This spot may contain cdc12p; alternatively, upon overexpression, cdc12p may accumulate at such a structure. (C) In early mitosis, the spot remains on the cortex. (D) In early mitosis, the ring forms at the site marked by the spot by extension of a strand from the spot around the circumference of the cell. (E) During anaphase, the ring becomes more robust. (F) After nuclear division, the ring closes during septation and cell division. Roles of the actin ring genes are inferred from cdc12p localization in mutants. Aspects of this model are inferred from cdc12p overexpression and mutant data. Alternate models are discussed in the text.
Similar articles
- The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin.
Kovar DR, Kuhn JR, Tichy AL, Pollard TD. Kovar DR, et al. J Cell Biol. 2003 Jun 9;161(5):875-87. doi: 10.1083/jcb.200211078. J Cell Biol. 2003. PMID: 12796476 Free PMC article. - The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis.
Balasubramanian MK, Hirani BR, Burke JD, Gould KL. Balasubramanian MK, et al. J Cell Biol. 1994 Jun;125(6):1289-301. doi: 10.1083/jcb.125.6.1289. J Cell Biol. 1994. PMID: 8207058 Free PMC article. - Movement of a cytokinesis factor cdc12p to the site of cell division.
Chang F. Chang F. Curr Biol. 1999 Jul 29-Aug 12;9(15):849-52. doi: 10.1016/s0960-9822(99)80372-8. Curr Biol. 1999. PMID: 10469572 - Microtubule and actin-dependent movement of the formin cdc12p in fission yeast.
Chang F. Chang F. Microsc Res Tech. 2000 Apr 15;49(2):161-7. doi: 10.1002/(SICI)1097-0029(20000415)49:2<161::AID-JEMT8>3.0.CO;2-2. Microsc Res Tech. 2000. PMID: 10816255 Review. - Regulation of contractile ring formation and septation in Schizosaccharomyces pombe.
Willet AH, McDonald NA, Gould KL. Willet AH, et al. Curr Opin Microbiol. 2015 Dec;28:46-52. doi: 10.1016/j.mib.2015.08.001. Epub 2015 Sep 3. Curr Opin Microbiol. 2015. PMID: 26340438 Free PMC article. Review.
Cited by
- A role for F-BAR protein Rga7p during cytokinesis in S. pombe.
Arasada R, Pollard TD. Arasada R, et al. J Cell Sci. 2015 Jul 1;128(13):2259-68. doi: 10.1242/jcs.162974. Epub 2015 May 14. J Cell Sci. 2015. PMID: 25977474 Free PMC article. - C-terminal anchoring of mid1p to membranes stabilizes cytokinetic ring position in early mitosis in fission yeast.
Celton-Morizur S, Bordes N, Fraisier V, Tran PT, Paoletti A. Celton-Morizur S, et al. Mol Cell Biol. 2004 Dec;24(24):10621-35. doi: 10.1128/MCB.24.24.10621-10635.2004. Mol Cell Biol. 2004. PMID: 15572668 Free PMC article. - The functionally distinct fission yeast formins have specific actin-assembly properties.
Scott BJ, Neidt EM, Kovar DR. Scott BJ, et al. Mol Biol Cell. 2011 Oct;22(20):3826-39. doi: 10.1091/mbc.E11-06-0492. Epub 2011 Aug 24. Mol Biol Cell. 2011. PMID: 21865598 Free PMC article. - Interaction of the N- and C-terminal autoregulatory domains of FRL2 does not inhibit FRL2 activity.
Vaillant DC, Copeland SJ, Davis C, Thurston SF, Abdennur N, Copeland JW. Vaillant DC, et al. J Biol Chem. 2008 Nov 28;283(48):33750-62. doi: 10.1074/jbc.M803156200. Epub 2008 Oct 2. J Biol Chem. 2008. PMID: 18835814 Free PMC article. - Progress towards understanding the mechanism of cytokinesis in fission yeast.
Pollard TD. Pollard TD. Biochem Soc Trans. 2008 Jun;36(Pt 3):425-30. doi: 10.1042/BST0360425. Biochem Soc Trans. 2008. PMID: 18481973 Free PMC article. Review.
References
- Balasubramanian MK, Helfman DM, Hemmingsen SM. A new tropomyosin essential for cytokinesis in the fission yeast S. pombe. . Nature (Lond) 1992;360:84–87. - PubMed
- Barbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. . Gene (Amst) 1992;114:59–66. - PubMed
- Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. - PubMed
- Castrillon DH, Wasserman SA. diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformitygene. Development (Camb) 1994;120:3367–3377. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials