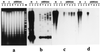Oat-maize chromosome addition lines: a new system for mapping the maize genome - PubMed (original) (raw)
Oat-maize chromosome addition lines: a new system for mapping the maize genome
E V Ananiev et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Novel plants with individual maize chromosomes added to a complete oat genome have been recovered via embryo rescue from oat (Avena sativa L., 2n = 6x = 42) x maize (Zea mays L., 2n = 20) crosses. An oat-maize disomic addition line possessing 21 pairs of oat chromosomes and one maize chromosome 9 pair was used to construct a cosmid library. A multiprobe (mixture of labeled fragments used as a probe) of highly repetitive maize-specific sequences was used to selectively isolate cosmid clones containing maize genomic DNA. Hybridization of individual maize cosmid clones or their subcloned fragments to maize and oat genomic DNA revealed that most high, middle, or low copy number DNA sequences are maize-specific. Such DNA markers allow the identification of maize genomic DNA in an oat genomic background. Chimeric cosmid clones were not found; apparently, significant exchanges of genetic material had not occurred between the maize-addition chromosome and the oat genome in these novel plants or in the cloning process. About 95% of clones selected at random from a maize genomic cosmid library could be detected by the multiprobe. The ability to selectively detect maize sequences in an oat background enables us to consider oat as a host for the cloning of specific maize chromosomes or maize chromosome segments. Introgressing maize chromosome segments into the oat genome via irradiation should allow the construction of a library of overlapping fragments for each maize chromosome to be used for developing a physical map of the maize genome.
Figures
Figure 1
Blot hybridization of a labeled multiprobe, composed of 22 maize-specific repetitive DNA sequences, to genomic DNA digests of 2 maize and 2 oat varieties. Strong signal is seen over lanes with maize DNA but not those with oat DNA. (a) EtdBr-stained 0.8% agarose gel after separation of DNA samples cut with _Eco_RI. (b) Autoradiogram after hybridization to the P-32 labeled multiprobe.
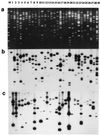
Figure 2
Blot panel of 29 maize-specific cosmid clones isolated from a cosmid library of an oat–maize chromosome 9 addition line. (a) Cosmids (–29) are cut by _Eco_RI restriction enzyme and size-fractionated in an 0.85% agarose gel stained with EtdBr (M = molecular weight marker, 1-kb ladder). (b) Labeled, total genomic maize DNA as a probe shows strong, medium, or weak hybridization to almost 90% of all _Eco_RI subfragments present in the cosmid clones. (c) Labeled multiprobe of highly repeated maize DNA sequences shows hybridization to from one to five _Eco_RI fragments in each lane.
Figure 3
Hybridization of unique and low-copy-number DNA sequences isolated from maize-specific cosmids shown in Fig. 2 to a blot panel of oat–maize chromosome addition lines carrying maize chromosomes 9, 7, 4, 3, and 2, respectively; m1 and m2 are maize stocks A188 and Seneca 60; o1 and o2 are oat stocks Sun II and Starter-1. (a) The 2.5-kb _Eco_RI fragment from cosmid 15 shows one band on the chromosome 9 addition line. An additional polymorphic band is present in the parental stocks of maize. (b) The 1.8-kb _Eco_RI fragment from cosmid 28 detects two bands. One band, polymorphic between m1 and m2, is present in the chromosome 9 addition line. An additional nonpolymorphic band is present on chromosome 9 and in both parental stocks. (c) The 2.1-kb _Eco_RI fragment from cosmid 10 shows one band on chromosome 9. (d) The 1.4-kb _Eco_RI fragment from cosmid 20 detects about 20 bands in parental maize stocks and several bands among the chromosome addition lines. The band pattern is chromosome-specific. No cross-hybridization occurred to oat DNA in a_–_d. (e) The 2.9-kb _Eco_RI fragment from cosmid 6 detects several polymorphic bands in parental maize stocks. One nonpolymorphic band is seen in maize, oat, and chromosomes 9, 7, 4, and 2 addition lines. Additional bands are seen in other chromosome addition lines.
Figure 4
Hybridization of medium and highly repetitive cloned maize DNA sequences to a blot panel of oat–maize chromosome addition lines carrying maize chromosomes 9, 7, 4, 3, and 2, respectively; m1 and m2 are maize stocks A188 and Seneca 60; o1 and o2 are oat stocks Sun II and Starter-1. (a) EtdBr-stained 0.8% agarose gel; lane M is a 1-kb molecular weight marker ladder. (b) The 0.7-kb _Eco_RI fragment of cosmid 10 shows multiple bands on different maize chromosomes as well as one common band for all chromosomes. (c) Cosmid 1, with a 40-kb insertion, shows strong hybridization signal over maize (lines m1 and m2) and over chromosome addition lines 9, 7, 4, 3, and 2; no hybridization is seen over oat DNA (lines o1 and o2). (d) With the 185-bp knob repeat, most of the hybridization signal is located on chromosome 9. No hybridization to oat DNA is detected.
Similar articles
- Addition of individual chromosomes of maize inbreds B73 and Mo17 to oat cultivars Starter and Sun II: maize chromosome retention, transmission, and plant phenotype.
Rines HW, Phillips RL, Kynast RG, Okagaki RJ, Galatowitsch MW, Huettl PA, Stec AO, Jacobs MS, Suresh J, Porter HL, Walch MD, Cabral CB. Rines HW, et al. Theor Appl Genet. 2009 Nov;119(7):1255-64. doi: 10.1007/s00122-009-1130-2. Epub 2009 Aug 26. Theor Appl Genet. 2009. PMID: 19707741 - A complete set of maize individual chromosome additions to the oat genome.
Kynast RG, Riera-Lizarazu O, Vales MI, Okagaki RJ, Maquieira SB, Chen G, Ananiev EV, Odland WE, Russell CD, Stec AO, Livingston SM, Zaia HA, Rines HW, Phillips RL. Kynast RG, et al. Plant Physiol. 2001 Mar;125(3):1216-27. doi: 10.1104/pp.125.3.1216. Plant Physiol. 2001. PMID: 11244103 Free PMC article. - Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions.
Ananiev EV, Phillips RL, Rines HW. Ananiev EV, et al. Proc Natl Acad Sci U S A. 1998 Oct 27;95(22):13073-8. doi: 10.1073/pnas.95.22.13073. Proc Natl Acad Sci U S A. 1998. PMID: 9789043 Free PMC article. - Complex structure of knobs and centromeric regions in maize chromosomes.
Ananiev EV, Phillips RL, Rines HW. Ananiev EV, et al. Tsitol Genet. 2000 Mar-Apr;34(2):11-5. Tsitol Genet. 2000. PMID: 10857197 Review. - Organization and variability of the maize genome.
Messing J, Dooner HK. Messing J, et al. Curr Opin Plant Biol. 2006 Apr;9(2):157-63. doi: 10.1016/j.pbi.2006.01.009. Epub 2006 Feb 3. Curr Opin Plant Biol. 2006. PMID: 16459130 Review.
Cited by
- Dissecting the maize genome by using chromosome addition and radiation hybrid lines.
Kynast RG, Okagaki RJ, Galatowitsch MW, Granath SR, Jacobs MS, Stec AO, Rines HW, Phillips RL. Kynast RG, et al. Proc Natl Acad Sci U S A. 2004 Jun 29;101(26):9921-6. doi: 10.1073/pnas.0403421101. Epub 2004 Jun 14. Proc Natl Acad Sci U S A. 2004. PMID: 15197265 Free PMC article. - Complex structure of knob DNA on maize chromosome 9. Retrotransposon invasion into heterochromatin.
Ananiev EV, Phillips RL, Rines HW. Ananiev EV, et al. Genetics. 1998 Aug;149(4):2025-37. doi: 10.1093/genetics/149.4.2025. Genetics. 1998. PMID: 9691055 Free PMC article. - Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic Loci shaped primarily by retrotransposons.
Wolfgruber TK, Sharma A, Schneider KL, Albert PS, Koo DH, Shi J, Gao Z, Han F, Lee H, Xu R, Allison J, Birchler JA, Jiang J, Dawe RK, Presting GG. Wolfgruber TK, et al. PLoS Genet. 2009 Nov;5(11):e1000743. doi: 10.1371/journal.pgen.1000743. Epub 2009 Nov 20. PLoS Genet. 2009. PMID: 19956743 Free PMC article. - The mechanisms and significance of the positional control of centromeres and telomeres in plants.
Oko Y, Ito N, Sakamoto T. Oko Y, et al. J Plant Res. 2020 Jul;133(4):471-478. doi: 10.1007/s10265-020-01202-2. Epub 2020 May 14. J Plant Res. 2020. PMID: 32410007 Review.
References
- Islam A K M R, Shepherd R, Sparrow D H B. Heredity. 1981;46:161–174.
- Riley R, Law C N. Stadler Genet Symp. 1984;16:301–322.
- Thomas H, Leggett J M, Jones I T. Euphytica. 1975;24:717–724.
- Jena K K, Khush G S. Genome. 1989;32:449–455.
- Jiang J, Morris K L, Gill B S. Chromosome Res. 1994;2:3–13. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources