Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1 - PubMed (original) (raw)
Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1
L A Hanakahi et al. Proc Natl Acad Sci U S A. 1997.
Abstract
LR1 is a B cell-specific, sequence-specific DNA binding activity that regulates transcription in activated B cells. LR1 also binds Ig heavy chain switch region sequences and may function in class switch recombination. LR1 contains two polypeptides, of 106 kDa and 45 kDa, and here we report that the 106-kDa component of LR1 is nucleolin. This identification, initially made by microsequence analysis, was verified by showing that (i) LR1-DNA binding activity increased in B cells transfected with a nucleolin cDNA expression construct; (ii) LR1-DNA binding activity was recognized by antibodies raised against recombinant human nucleolin; and (iii) in B cells transfected with epitope-tagged nucleolin expression constructs, the LR1-DNA complex was recognized by the anti-tag antibody. Nucleolin is an abundant nucleolar protein which is believed to play a role in rDNA transcription or organization, or rRNA processing. Homology between nucleolin and histone H1 suggests that nucleolin may alter DNA organization in response to cell cycle controls, and the nucleolin component of LR1 may therefore function to organize switch regions before, during, or after switch recombination. The demonstration that nucleolin is a component of a B cell-specific complex that binds switch region sequences suggests that the G-rich switch regions may have evolved from rDNA.
Figures
Figure 1
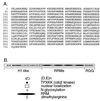
Nucleolin. (A) Predicted amino acid sequence of human nucleolin. Residues 624–626 were omitted from the original published report (GenBank file humnucleo, accession number J05584J05584; ref. 22). (B) Schematic of nucleolin, showing the histone H1-like N-terminal region, which includes long runs of acidic amino acids and sites for cdc2 kinase; the nuclear localization signal and N-glycosylation sites; the four RRMs; and the RGG motifs in the C terminus.
Figure 2
Specificity of antinucleolin antibodies. (A) In situ immunofluorescent staining of PD31 murine pre-B cells with antinucleolin antibodies. Arrows indicate two of the nucleoli evident by staining. (B) Immunoblot analysis of crude PD31 nuclear extract; the single band at 106 kDa is indicated.
Figure 3
Gel mobility-shift analysis of the effect of antinucleolin antibodies on LR1–DNA binding activity. (A) Nuclear extract from PD31 pre-B cells was treated with no antibodies (no Ab) or 2 μg of protein A-purified antibodies from pre-immune serum (pre) or from a rabbit immunized with recombinant human nucleolin (α-nuc). Arrows indicate bands corresponding to the LR1–DNA complex (LR1), the subshift, and free DNA duplex. (B) LR1 purified 12,000-fold from PD31 pre-B cells was treated with 0, 0.016, 0.08, 0.4 or 2 μg of protein A purified antibodies from pre-immune serum (pre) or from a rabbit immunized with recombinant human nucleolin (α-nuc). Control lanes on the right show that neither antibody preparation alone altered mobility of the DNA duplex. (C) Purified LR1 was treated with 2 μg rabbit polyclonal antinucleolin antibodies in the presence of antirabbit Ig (α-rIg) or antirabbit IgM antibodies (α-rIgM) at 1 or 5 μg/reaction.
Figure 4
Epitope-tagged nucleolin is found in the LR1–DNA binding complex. PD31 pre-B cells were mock-transfected (−) or transfected with the constructs indicated. (A) Gel mobility-shift analysis of nuclear extracts of PD31 pre-B cells transfected with increasing amounts (1, 2, 4, or 16 μg) of pNtag4, which expresses nucleolin cDNA carrying an N-terminal HA tag. (B) Gel mobility-shift analysis of nuclear extracts from cells transfected with 4 μg pNtag4, which expresses nucleolin cDNA carrying an N-terminal HA tag; or 4 μg pNfor4, which expresses untagged nucleolin cDNA. Reactions were treated with 0.3 or 1 μl of anti-tag mAb 12CA5, as indicated. Control lanes on the right show that the 12CA5 antibody preparation alone did not alter the mobility of the DNA duplex.
Figure 5
Gel mobility-shift analysis of LR1 binding to sites substituted with inosine (I) and 2-aminopurine (2-AP). Binding specificity of LR1 in PD31 nuclear extracts is compared in the absence (Left) and presence (Right) of antinucleolin antibodies.
Similar articles
- Transcriptional activation by LR1 at the Emu enhancer and switch region sites.
Hanakahi LA, Maizels N. Hanakahi LA, et al. Nucleic Acids Res. 2000 Jul 15;28(14):2651-7. doi: 10.1093/nar/28.14.2651. Nucleic Acids Res. 2000. PMID: 10908319 Free PMC article. - G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D, A role for G-G pairing in immunoglobulin switch recombination.
Dempsey LA, Sun H, Hanakahi LA, Maizels N. Dempsey LA, et al. J Biol Chem. 1999 Jan 8;274(2):1066-71. doi: 10.1074/jbc.274.2.1066. J Biol Chem. 1999. PMID: 9873052 - The ubiquitously expressed DNA-binding protein late SV40 factor binds Ig switch regions and represses class switching to IgA.
Drouin EE, Schrader CE, Stavnezer J, Hansen U. Drouin EE, et al. J Immunol. 2002 Mar 15;168(6):2847-56. doi: 10.4049/jimmunol.168.6.2847. J Immunol. 2002. PMID: 11884454 - Molecular dissection of nucleolin's role in growth and cell proliferation: new insights.
Srivastava M, Pollard HB. Srivastava M, et al. FASEB J. 1999 Nov;13(14):1911-22. FASEB J. 1999. PMID: 10544174 Review. - Molecular mechanism of immunoglobulin V-region diversification regulated by transcription and RNA metabolism in antigen-driven B cells.
Sakaguchi N, Maeda K, Kuwahara K. Sakaguchi N, et al. Scand J Immunol. 2011 Jun;73(6):520-6. doi: 10.1111/j.1365-3083.2011.02557.x. Scand J Immunol. 2011. PMID: 21388430 Review.
Cited by
- A nucleolin-binding 3' untranslated region element stabilizes beta-globin mRNA in vivo.
Jiang Y, Xu XS, Russell JE. Jiang Y, et al. Mol Cell Biol. 2006 Mar;26(6):2419-29. doi: 10.1128/MCB.26.6.2419-2429.2006. Mol Cell Biol. 2006. PMID: 16508016 Free PMC article. - A human nuclease specific for G4 DNA.
Sun H, Yabuki A, Maizels N. Sun H, et al. Proc Natl Acad Sci U S A. 2001 Oct 23;98(22):12444-9. doi: 10.1073/pnas.231479198. Proc Natl Acad Sci U S A. 2001. PMID: 11675489 Free PMC article. - Transcriptional activation by LR1 at the Emu enhancer and switch region sites.
Hanakahi LA, Maizels N. Hanakahi LA, et al. Nucleic Acids Res. 2000 Jul 15;28(14):2651-7. doi: 10.1093/nar/28.14.2651. Nucleic Acids Res. 2000. PMID: 10908319 Free PMC article. - Nucleolin is important for Epstein-Barr virus nuclear antigen 1-mediated episome binding, maintenance, and transcription.
Chen YL, Liu CD, Cheng CP, Zhao B, Hsu HJ, Shen CL, Chiu SJ, Kieff E, Peng CW. Chen YL, et al. Proc Natl Acad Sci U S A. 2014 Jan 7;111(1):243-8. doi: 10.1073/pnas.1321800111. Epub 2013 Dec 16. Proc Natl Acad Sci U S A. 2014. PMID: 24344309 Free PMC article. - Nucleolin stabilizes Bcl-X L messenger RNA in response to UVA irradiation.
Zhang J, Tsaprailis G, Bowden GT. Zhang J, et al. Cancer Res. 2008 Feb 15;68(4):1046-54. doi: 10.1158/0008-5472.CAN-07-1927. Cancer Res. 2008. PMID: 18281479 Free PMC article.
References
- Williams M, Maizels N. Genes Dev. 1991;5:2353–2361. - PubMed
- Williams M, Hanakahi L A, Maizels N. J Biol Chem. 1993;268:13731–13737. - PubMed
- Mowatt M R, Dunnick W A. J Immunol. 1986;136:2674–2683. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA16038/CA/NCI NIH HHS/United States
- R01 GM39799/GM/NIGMS NIH HHS/United States
- P01 CA016038/CA/NCI NIH HHS/United States
- R01 GM039799/GM/NIGMS NIH HHS/United States
- F32 GM015948/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources