Design of TATA box-binding protein/zinc finger fusions for targeted regulation of gene expression - PubMed (original) (raw)
Design of TATA box-binding protein/zinc finger fusions for targeted regulation of gene expression
J S Kim et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Fusing the TATA box-binding protein (TBP) to other DNA-binding domains may provide a powerful way of targeting TBP to particular promoters. To explore this possibility, a structure-based design strategy was used to construct a fusion protein, TBP/ZF, in which the three zinc fingers of Zif268 were linked to the COOH terminus of yeast TBP. Gel shift experiments revealed that this fusion protein formed an extraordinarily stable complex when bound to the appropriate composite DNA site (half-life up to 630 h). In vitro transcription experiments and transient cotransfection assays revealed that TBP/ZF could act as a site-specific repressor. Because the DNA-binding specificities of zinc finger domains can be systematically altered by phage display, it may be possible to target such TBP/zinc finger fusions to desired promoters and thus specifically regulate expression of endogenous genes.
Figures
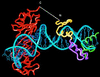
Figure 1
Structure-based design of TBP/ZF. The cocrystal structures of the Zif268:DNA (14) and the TBP:TATA box complexes (9) were aligned by superimposing phosphates in several different registers. In the model shown above, the NH2-terminal end of Zif268 was 23 Å away from the COOH-terminal end of TBP. We created the TBP/ZF fusion protein by adding a NH2-(Gly-Gly-Gly-Ser)2Gly-COOH polypeptide linker to join the two molecules. The alignment of binding sites used in this modeling study suggested that TBP/ZF would bind tightly to the sequence 5′-GCGTGGGCGNNNNTATATAAA-3′.
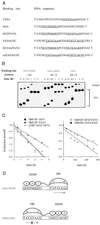
Figure 2
Determination of dissociation rate constants. (A) Probe DNA sequences used in gel shift assays. The TATA boxes and the Zif268-binding site are underlined. The sequence of only one strand is shown. (B) Example of gel shift assay used to determine dissociation rate constants. yTBP (5 μg/ml) or TBP/ZF (6 μg/ml) was incubated with labeled probe DNAs (0.1 nM) for 1 h at room temperature. To begin measurement of dissociation rates, a large excess of unlabeled probe DNA (final concentration, 1 μM) was added to each incubation mixture at time t = 0. Aliquots were removed at indicated times and analyzed by gel electrophoresis. Samples were loaded on the gel at different times, and thus the bands appear staggered. (C) The fraction of labeled probe DNA bound by protein was quantified by PhosphorImager (Molecular Dynamics) analysis, and normalized to the fraction bound at time t = 0. The natural log of the normalized fraction bound was plotted against time, and the dissociation rate was determined from the slope. (D) Models indicating how the orientationof the TBP moiety of TBP/ZF on the TATA box may be controlled by flanking Zif268-binding sites. The direction of transcription relative to the TATA box is shown with an arrow. (The “x” over the lower arrow indicates that this TBP orientation cannot support transcription.)
Figure 3
In vitro transcription analysis. Biotinylated DNA fragments containing the promoters (TATA, TATA/CGC, and GCG/TATA) upstream of a G-free cassette were immobilized on streptavidin-coupled paramagnetic beads and used as transcription templates. yTBP at 6 μg/ml (lanes 1–3 and 7–9) or TBP/ZF at 8 μg/ml (lanes 4–6 and 10–12) was preincubated with each template (0.1 nM) for 1 h at room temperature. Then, supernatants were removed, and excess amounts (1 μM each) of competitor DNA oligonucleotides (GCG and TATA from Fig. 2_A_) were added to the preincubation mixture. After incubating 24 h at 4°C, the beads were washed to remove proteins that dissociated from the templates, and human transcription factors (TFIIB, -IIE, -IIF, and -IIH), RNA polymerase II, and substrate nucleotides were added to initiate transcription. yTBP was also added to a final concentration of 0.2 μg/ml in lanes 7–12. The transcripts were analyzed by urea gel electrophoresis.
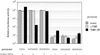
Figure 4
Transient cotransfection assay. Human 293 cells were cotransfected, using the calcium phosphate precipitation method with (i) 1 μg of expression plasmid encoding yTBP or TBP/ZF, (ii) 5 μg of activator plasmid, GAL4-VP16, (iii) 0.5 μg of β-galactosidase expression plasmid (pCMVβ) as an internal control, (iv) 1 μg of a reporter plasmid (derived from pGL3-Basic) encoding the firefly luciferase gene, and (v) variable amount of the carrier plasmid (pUC19) to keep the total amount of transfected DNA at 20 μg. Each reporter construct had five GAL4-binding sites upstream of one of the promoter sequences (TATA, TATA/CGC, or GCG/TATA) used in the in vitro transcription assay (Fig. 3). In a parallel assay of basal transcription, GAL4-VP16 was omitted. Luciferase activity was measured 2 days after transfection and was normalized (i) with respect to β-galactosidase activity (to correct for transfection efficiency), and (ii) to the corresponding value from the cells transfected with blank expression vector, pcDNA3 (which was set to an arbitrary value of 104). The absence or presence of GAL4-VP16 is indicated. The data represent an average of three independent experiments, and the standard error of the mean is shown.
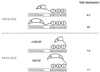
Figure 5
Transcriptional repression by TBP/ZF and ΔTBP/ZF in vivo. Transient cotransfection assays were used to determine whether TBP/ZF and ΔTBP/ZF could affect VP16-activated transcription from the TATA/GCG promoter. The results were compared with those from the TATA/CGC promoter (Fig. 4). The data represent an average of three independent experiments.
Similar articles
- Structure-based design of a dimeric zinc finger protein.
Pomerantz JL, Wolfe SA, Pabo CO. Pomerantz JL, et al. Biochemistry. 1998 Jan 27;37(4):965-70. doi: 10.1021/bi972464o. Biochemistry. 1998. PMID: 9467467 - Combining structure-based design with phage display to create new Cys(2)His(2) zinc finger dimers.
Wolfe SA, Ramm EI, Pabo CO. Wolfe SA, et al. Structure. 2000 Jul 15;8(7):739-50. doi: 10.1016/s0969-2126(00)00161-1. Structure. 2000. PMID: 10903945 - Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code.
Wolfe SA, Greisman HA, Ramm EI, Pabo CO. Wolfe SA, et al. J Mol Biol. 1999 Feb 5;285(5):1917-34. doi: 10.1006/jmbi.1998.2421. J Mol Biol. 1999. PMID: 9925775 - Mechanisms of transcriptional activation and repression can both involve TFIID.
Manley JL, Um M, Li C, Ashali H. Manley JL, et al. Philos Trans R Soc Lond B Biol Sci. 1996 Apr 29;351(1339):517-26. doi: 10.1098/rstb.1996.0050. Philos Trans R Soc Lond B Biol Sci. 1996. PMID: 8735274 Review. - Designer zinc-finger proteins and their applications.
Papworth M, Kolasinska P, Minczuk M. Papworth M, et al. Gene. 2006 Jan 17;366(1):27-38. doi: 10.1016/j.gene.2005.09.011. Epub 2005 Nov 17. Gene. 2006. PMID: 16298089 Review.
Cited by
- A novel four zinc-finger protein targeted against p190(BcrAbl) fusion oncogene cDNA: utilisation of zinc-finger recognition codes.
McNamara AR, Ford KG. McNamara AR, et al. Nucleic Acids Res. 2000 Dec 15;28(24):4865-72. doi: 10.1093/nar/28.24.4865. Nucleic Acids Res. 2000. PMID: 11121477 Free PMC article. - Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites.
Tan W, Zhu K, Segal DJ, Barbas CF 3rd, Chow SA. Tan W, et al. J Virol. 2004 Feb;78(3):1301-13. doi: 10.1128/jvi.78.3.1301-1313.2004. J Virol. 2004. PMID: 14722285 Free PMC article. - Therapeutic modulation of endogenous gene function by agents with designed DNA-sequence specificities.
Uil TG, Haisma HJ, Rots MG. Uil TG, et al. Nucleic Acids Res. 2003 Nov 1;31(21):6064-78. doi: 10.1093/nar/gkg815. Nucleic Acids Res. 2003. PMID: 14576293 Free PMC article. Review. - Characterisation of site-biased DNA methyltransferases: specificity, affinity and subsite relationships.
McNamara AR, Hurd PJ, Smith AE, Ford KG. McNamara AR, et al. Nucleic Acids Res. 2002 Sep 1;30(17):3818-30. doi: 10.1093/nar/gkf501. Nucleic Acids Res. 2002. PMID: 12202767 Free PMC article. - Improved xylose tolerance and 2,3-butanediol production of Klebsiella pneumoniae by directed evolution of rpoD and the mechanisms revealed by transcriptomics.
Guo XW, Zhang Y, Li LL, Guan XY, Guo J, Wu DG, Chen YF, Xiao DG. Guo XW, et al. Biotechnol Biofuels. 2018 Nov 9;11:307. doi: 10.1186/s13068-018-1312-8. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 30455736 Free PMC article.
References
- Buratowski S, Hahn S, Guarente L, Sharp P A. Cell. 1989;56:549–561. - PubMed
- Klein C, Struhl K. Science. 1994;266:280–282. - PubMed
- Colgan J, Manley J L. Genes Dev. 1992;6:304–315. - PubMed
- Stringer K F, Ingles C J, Greenblatt J. Nature (London) 1990;345:783–786. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources