The E6-Ap ubiquitin-protein ligase (UBE3A) gene is localized within a narrowed Angelman syndrome critical region - PubMed (original) (raw)
- PMID: 9110176
- PMCID: PMC139148
- DOI: 10.1101/gr.7.4.368
The E6-Ap ubiquitin-protein ligase (UBE3A) gene is localized within a narrowed Angelman syndrome critical region
J S Sutcliffe et al. Genome Res. 1997 Apr.
Abstract
Angelman syndrome (AS) and Prader-Willi syndrome (PWS) are distinct clinical phenotypes resulting from maternal and paternal deficiencies, respectively, in human chromosome 15qll-q13. Although several imprinted, paternally expressed transcripts have been identified within the PWS candidate region, no maternally expressed gene has yet been identified within the AS candidate region. We have developed an integrated physical map spanning the PWS and AS candidate regions and localized two breakpoints, including a cryptic t(14;15) translocation associated with AS and a non-AS 15q deletion, which substantially narrow the AS candidate region to approximately 250 kb. Mapping data indicate that the entire transcriptional unit of the E6-AP ubiquitin-protein ligase (UBE3A) gene lies within the AS region. The UBE3A locus expresses a transcript of approximately 5 kb at low to moderate levels in all tissues tested. The mouse homolog of UBE3A was cloned and sequenced revealing a high degree of conservation at nucleotide and protein levels. Northern and RT-PCR analysis of Ube3a expression in mouse tissues from animals with segmental, paternal uniparental disomy failed to detect substantially reduced or absent expression compared to control animals, failing to provide any evidence for maternal-specific expression from this locus. Recent identification of de novo truncating mutations in UBE3A taken with these observations indicates that mutations in UBE3A can lead to AS and suggests that this locus may encode both imprinted and biallelically expressed products.
Figures
Figure 1
(A) A schematic physical map of the 15q11–q13 PWS/AS common deletion interval is shown with the centromere toward the left and the telomere toward the right. Genes and genomic markers are shown in boxes. Sites of differential methylation are indicated by asterisks (*) over PW71 and SNRPN. Wavy vertical lines represent chromosomal breakpoints. The common PWS and AS deletion breakpoints are near either end of the map, and the Se family centromeric deletion breakpoint maps between SRRPN and UBE3A, defining the PWS and AS candidate regions, indicated over the map. Breakpoints in the 15q non-AS deletion case and the t(14;15) translocation case, together with the Se breakpoint, define the narrowed AS critical region indicated above UBE3A. (B) YAC, PAC, cosmid, STS, and gene map of the ∼1-Mb region surrounding the Se breakpoint. Genomic clones are indicated by horizontal lines; gene and STS markers are indicated by broken vertical lines.
Figure 2
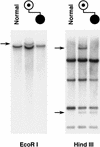
Detection of the t(14;15) cryptic translocation breakpoints. An 8-kb _Eco_RI fragment from cosmid 24 detects a single translocation junction fragment in _Eco_RI-digested DNA from the mother but not from the child, who is unbalanced. Both der(14) and der(15) breakpoints are seen in the _Hin_dIII panel, but the unbalanced, affected child has only the der(14) and not the der(15) chromosome, therefore displaying only one of the two breakpoint fragments seen in the mother.
Figure 3
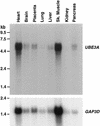
Northern analysis of UBE3A gene expression. Hybridization of UBE3A and control GAP3D cDNA probes to a Northern filter (Clontech) containing 2 μg poly(A)+ RNA per lane is shown.
Figure 4
Comparison of peptide sequences for human and mouse UBE3A. The deduced peptide sequence of the mouse cDNA was compared to the published human peptide sequence, using the Wisconsin package; this analysis revealed 94% amino acid identity between human and mouse for UBE3A.
Figure 5
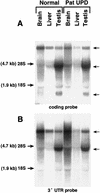
Northern analysis of Ube3a gene expression in mouse tissues from control and paternal UPD mice using probes corresponding to the coding region (A) and the 3′ UTR region (B). Comparison to the ethidium bromide-stained gel (not shown) reveals no significant difference between the control and UPD animals.
Similar articles
- De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome.
Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. Matsuura T, et al. Nat Genet. 1997 Jan;15(1):74-7. doi: 10.1038/ng0197-74. Nat Genet. 1997. PMID: 8988172 - Imprinting in Angelman and Prader-Willi syndromes.
Jiang Y, Tsai TF, Bressler J, Beaudet AL. Jiang Y, et al. Curr Opin Genet Dev. 1998 Jun;8(3):334-42. doi: 10.1016/s0959-437x(98)80091-9. Curr Opin Genet Dev. 1998. PMID: 9691003 Review. - The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A.
Runte M, Hüttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. Runte M, et al. Hum Mol Genet. 2001 Nov 1;10(23):2687-700. doi: 10.1093/hmg/10.23.2687. Hum Mol Genet. 2001. PMID: 11726556 - Genomic imprinting: potential function and mechanisms revealed by the Prader-Willi and Angelman syndromes.
Glenn CC, Driscoll DJ, Yang TP, Nicholls RD. Glenn CC, et al. Mol Hum Reprod. 1997 Apr;3(4):321-32. doi: 10.1093/molehr/3.4.321. Mol Hum Reprod. 1997. PMID: 9237260 Review. - Prader-Willi syndrome and Angelman syndrome.
Buiting K. Buiting K. Am J Med Genet C Semin Med Genet. 2010 Aug 15;154C(3):365-76. doi: 10.1002/ajmg.c.30273. Am J Med Genet C Semin Med Genet. 2010. PMID: 20803659 Review.
Cited by
- Alterations in intrinsic membrane properties and the axon initial segment in a mouse model of Angelman syndrome.
Kaphzan H, Buffington SA, Jung JI, Rasband MN, Klann E. Kaphzan H, et al. J Neurosci. 2011 Nov 30;31(48):17637-48. doi: 10.1523/JNEUROSCI.4162-11.2011. J Neurosci. 2011. PMID: 22131424 Free PMC article. - Ube3a imprinting impairs circadian robustness in Angelman syndrome models.
Shi SQ, Bichell TJ, Ihrie RA, Johnson CH. Shi SQ, et al. Curr Biol. 2015 Mar 2;25(5):537-45. doi: 10.1016/j.cub.2014.12.047. Epub 2015 Feb 5. Curr Biol. 2015. PMID: 25660546 Free PMC article. - A ubiquitin variant-based affinity approach selectively identifies substrates of the ubiquitin ligase E6AP in complex with HPV-11 E6 or HPV-16 E6.
Ebner FA, Sailer C, Eichbichler D, Jansen J, Sladewska-Marquardt A, Stengel F, Scheffner M. Ebner FA, et al. J Biol Chem. 2020 Oct 30;295(44):15070-15082. doi: 10.1074/jbc.RA120.015603. Epub 2020 Aug 27. J Biol Chem. 2020. PMID: 32855237 Free PMC article. - Molecular characterisation of four cases of intrachromosomal triplication of chromosome 15q11-q14.
Ungaro P, Christian SL, Fantes JA, Mutirangura A, Black S, Reynolds J, Malcolm S, Dobyns WB, Ledbetter DH. Ungaro P, et al. J Med Genet. 2001 Jan;38(1):26-34. doi: 10.1136/jmg.38.1.26. J Med Genet. 2001. PMID: 11134237 Free PMC article. - Allelic specificity of Ube3a expression in the mouse brain during postnatal development.
Judson MC, Sosa-Pagan JO, Del Cid WA, Han JE, Philpot BD. Judson MC, et al. J Comp Neurol. 2014 Jun 1;522(8):1874-96. doi: 10.1002/cne.23507. J Comp Neurol. 2014. PMID: 24254964 Free PMC article.
References
- Buiting K, Dittrich B, Gross S, Greger V, Lalande M, Robinson W, Mutirangura A, Ledbetter DH, Horsthemke B. Molecular definition of the Prader-Willi syndrome chromosome region and orientation of the SNRPN gene. Hum Mol Genet. 1993;2:1991–1994. - PubMed
- Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls RD, Horsthemke B. Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nature Genet. 1995;9:395–400. - PubMed
- Buxion JL, Chan CT, Gilbert H, Clayton-Smith J, Burn J, Pembrey M, Malcolm S. Angelman syndrome associated with a maternal 15q11-13 deletion of less than 200 kb. Hum Mol Genet. 1994;3:1409–1413. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials