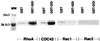RhoGDIgamma: a GDP-dissociation inhibitor for Rho proteins with preferential expression in brain and pancreas - PubMed (original) (raw)
Comparative Study
RhoGDIgamma: a GDP-dissociation inhibitor for Rho proteins with preferential expression in brain and pancreas
C N Adra et al. Proc Natl Acad Sci U S A. 1997.
Abstract
GDP-dissociation inhibitors (GDIs) play a primary role in modulating the activation of GTPases and may also be critical for the cellular compartmentalization of GTPases. RhoGDI and GDI/D4 are two currently known GDIs for the Rho-subfamily of GTPases. Using their cDNAs to screen a human brain cDNA library under low stringency, we have cloned a homologous cDNA preferentially expressed at high levels in brain and pancreas. The predicted protein, named RhoGDIgamma, is approximately 50% identical to GDI/D4 and RhoGDI. It binds to CDC42 and RhoA with less affinity compared with RhoGDI and does not bind with Rac1, Rac2, or Ras. RhoGDIgamma functions as a GDI for CDC42 but with approximately 20 times less efficiency than RhoGDI. Immunohistochemical studies showed a diffuse punctate distribution of the protein in the cytoplasm with concentration around the nucleus in cytoplasmic vesicles. Overexpression of the protein in baby hamster kidney cells caused the cells to round up with loss of stress fibers. A distinct hydrophobic amino terminus in RhoGDIgamma, not seen in the other two RhoGDIs, could provide a mechanism for localization of the GDI to specific membranous compartment thus determining function distinct from RhoGDI or GDI/D4. Our results provide evidence that there is a family of GDIs for the Rho-related GTPases and that they differ in binding affinity, target specificity, and tissue expression. We propose that RhoGDI be renamed RhoGDIalpha and GDID4 be renamed RhoGDIbeta. The new GDI should widen the scope of investigation of this important class of regulatory protein.
Figures
Figure 1
Nucleotide and predicted amino acid sequence of RhoGDIγ. Nucleotides are numbered together with amino acid sequence of the longest ORF numbered from the presumed initiating methionine. A TGA stop codon (∗∗∗) is followed by a short 3′ untranslated region.
Figure 2
Comparison of amino acid sequence of RhoGDIγ with RhoGDI (RhoGDIα) and GDI/D4 (RhoGDIβ). Identical residues are indicated by straight lines, conserved residues are indicated by ∗. Conservative amino acids are grouped as follows: S, T, G, A, P; L, M, I, V; E, D, Q, N; R, K, H; F, Y, W; C. The hydrophobic domain in the amino-terminal region of RhoGDIγ is underlined.
Figure 3
Detection of RhoGDIγ mRNA in poly(A)+ RNA from normal human tissues. The filter (CLONTECH) contained 2 μg of poly(A)+ RNA per lane. The filter was hybridized with 32P-labeled RhoGDIγ cDNA probe, and washed with 0.2× standard saline citrate at 65°C before autoradiography for 24 hr at −80°C. Positions for 4.4 kb and 1.35 kb as marked by ribosomal markers are indicated.
Figure 4
Binding of RhoGDIγ to RhoA, CDC42, Rac1, and Rac2. Rho proteins were produced in insect cells infected with the various baculovirus as described in Materials and Methods. GST–RhoGDIγ bound to glutathione agarose beads was then incubated with lysates of insects cells for 18 hr at 4°C. GST alone was used as control. Bound proteins were separated by SDS/PAGE and Western filters were then probed with the respective antibodies (Santa Cruz Biotechnology) against RhoA, CDC42, Rac1, or Rac2. The figure shows binding of RhoA and CDC42 to RhoGDIγ (GST–GDI). The binding of Rac1 or Rac2 by GST–RhoGDIγ was the same as background binding by GST.
Figure 5
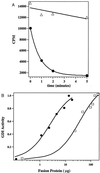
Inhibition of GDP-dissociation from CDC42Hs by RhoGDIγ. (A) Time course of [3H]GDP dissociation from CDC42 upon mixing with low Mg2+ (7.5 mM EDTA) buffer in the presence of RhoGDIγ (▵) or control buffer (•). (B) Comparison of the relative efficiency of RhoGDI (•) and RhoGDIγ(□) in eliciting the GDI effect. CDC42Hs-[3H]GDP was incubated with the indicated amount of GDI, and the amount of radioactivity associated with CDC42Hs 5 min after initiating dissociation was determined by scintillation counter. The amount of the [3H]GDP still bound to the CDC42Hs protein was then normalized as a percentage of the amount bound in the presence of the maximum concentration of RhoGDI. The figure shows that the activity of RhoGDIγ is ≈20-fold less than that of RhoGDI.
Figure 6
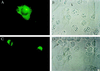
Immunolocalization of RhoGDIγ. Plasmid DNA of GFP-tagged RhoGDIγ vector was transfected into baby hamster kidney cells grown on coverslips using the calcium phosphate precipitate method. At different times after transfection, the cells were fixed and the coverslips mounted in glycerol. Cells were examined under Argon arc light activation. (A) A cell expressing RhoGDIγ 24 hr after transfection, showing the typical diffuse punctate distribution of the protein with concentration in the perinuclear region. The yellowish spots are due to unavoidable overexposure from intensely fluorescent locations. (B) Same field as A taken under phase contrast. The positive cell is indicated by an arrow. (C) Two cells expressing very high levels of RhoGDIγ. (D) Same field as C under phase contrast, showing that the positive cells (arrows) are rounded up compared with neighboring normal adherent baby hamster kidney cells.
Similar articles
- RhoGDI-3 is a new GDP dissociation inhibitor (GDI). Identification of a non-cytosolic GDI protein interacting with the small GTP-binding proteins RhoB and RhoG.
Zalcman G, Closson V, Camonis J, Honoré N, Rousseau-Merck MF, Tavitian A, Olofsson B. Zalcman G, et al. J Biol Chem. 1996 Nov 29;271(48):30366-74. doi: 10.1074/jbc.271.48.30366. J Biol Chem. 1996. PMID: 8939998 - Identification of a novel protein with GDP dissociation inhibitor activity for the ras-like proteins CDC42Hs and rac I.
Adra CN, Ko J, Leonard D, Wirth LJ, Cerione RA, Lim B. Adra CN, et al. Genes Chromosomes Cancer. 1993 Dec;8(4):253-61. doi: 10.1002/gcc.2870080408. Genes Chromosomes Cancer. 1993. PMID: 7512369 - The identification and characterization of a GDP-dissociation inhibitor (GDI) for the CDC42Hs protein.
Leonard D, Hart MJ, Platko JV, Eva A, Henzel W, Evans T, Cerione RA. Leonard D, et al. J Biol Chem. 1992 Nov 15;267(32):22860-8. J Biol Chem. 1992. PMID: 1429634 - Rho guanine dissociation inhibitors: pivotal molecules in cellular signalling.
Olofsson B. Olofsson B. Cell Signal. 1999 Aug;11(8):545-54. doi: 10.1016/s0898-6568(98)00063-1. Cell Signal. 1999. PMID: 10433515 Review. - Underappreciated roles for Rho GDP dissociation inhibitors (RhoGDIs) in cell function: Lessons learned from the pancreatic islet β-cell.
Kowluru A, Gleason NF. Kowluru A, et al. Biochem Pharmacol. 2022 Mar;197:114886. doi: 10.1016/j.bcp.2021.114886. Epub 2021 Dec 28. Biochem Pharmacol. 2022. PMID: 34968495 Free PMC article. Review.
Cited by
- Identification and characterisation of the RalA-ERp57 interaction: evidence for GDI activity of ERp57.
Brymora A, Duggin IG, Berven LA, van Dam EM, Roufogalis BD, Robinson PJ. Brymora A, et al. PLoS One. 2012;7(11):e50879. doi: 10.1371/journal.pone.0050879. Epub 2012 Nov 30. PLoS One. 2012. PMID: 23226417 Free PMC article. - Cholecystokinin-mediated RhoGDI phosphorylation via PKCα promotes both RhoA and Rac1 signaling.
Sabbatini ME, Williams JA. Sabbatini ME, et al. PLoS One. 2013 Jun 11;8(6):e66029. doi: 10.1371/journal.pone.0066029. Print 2013. PLoS One. 2013. PMID: 23776598 Free PMC article. - Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding.
Michaelson D, Silletti J, Murphy G, D'Eustachio P, Rush M, Philips MR. Michaelson D, et al. J Cell Biol. 2001 Jan 8;152(1):111-26. doi: 10.1083/jcb.152.1.111. J Cell Biol. 2001. PMID: 11149925 Free PMC article. - The hematopoiesis-specific GTP-binding protein RhoH is GTPase deficient and modulates activities of other Rho GTPases by an inhibitory function.
Li X, Bu X, Lu B, Avraham H, Flavell RA, Lim B. Li X, et al. Mol Cell Biol. 2002 Feb;22(4):1158-71. doi: 10.1128/MCB.22.4.1158-1171.2002. Mol Cell Biol. 2002. PMID: 11809807 Free PMC article. - Cullin 3/KCTD5 Promotes the Ubiqutination of Rho Guanine Nucleotide Dissociation Inhibitor 1 and Regulates Its Stability.
Cho HJ, Ryu KJ, Baek KE, Lim J, Kim T, Song CY, Yoo J, Lee HG. Cho HJ, et al. J Microbiol Biotechnol. 2020 Oct 28;30(10):1488-1494. doi: 10.4014/jmb.2007.07033. J Microbiol Biotechnol. 2020. PMID: 32876072 Free PMC article.
References
- Boguski M S, McCormick F. Nature (London) 1993;366:643–654. - PubMed
- Fukumoto Y, Kaibuchi K, Hori Y, Fujiko H, Araki S, Ueda T, Kikuchi A, Takai Y. Oncogene. 1990;5:1321–1328. - PubMed
- Leonard D, Hart M, Platko J, Eva A, Henzel W, Evans T, Cerione R A. J Biol Chem. 1992;267:22860–22868. - PubMed
- Abo A, Pick E, Hall A, Totty N, Teahan C, Segal A. Nature (London) 1991;353:668–670. - PubMed
- Knaus U, Keyworth P, Kinsella B, Curnette J, Bokoch G M. J Biol Chem. 1992;267:23575–23582. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous