Mutations in mitochondrial cytochrome c oxidase genes segregate with late-onset Alzheimer disease - PubMed (original) (raw)
Comparative Study
Mutations in mitochondrial cytochrome c oxidase genes segregate with late-onset Alzheimer disease
R E Davis et al. Proc Natl Acad Sci U S A. 1997.
Retraction in
- Retraction.
Davis RE, Miller S, Herrnstadt C, Ghosh SS, Fahy E, Shinobu LA, Galasko D, Thal LJ, Beal MF, Howell N, Parker WD. Davis RE, et al. Proc Natl Acad Sci U S A. 1998 Sep 29;95(20):12069. doi: 10.1073/pnas.95.20.12069-b. Proc Natl Acad Sci U S A. 1998. PMID: 16578857 Free PMC article. No abstract available.
Abstract
Mounting evidence suggests that defects in energy metabolism contribute to the pathogenesis of Alzheimer disease (AD). Cytochrome c oxidase (CO) is kinetically abnormal, and its activity is decreased in brain and peripheral tissue in late-onset AD. CO is encoded by both the mitochondrial and the nuclear genomes. Its catalytic centers, however, are encoded exclusively by two mitochondrial genes, CO1 and CO2 (encoding CO subunits I and II, respectively). We searched these genes, as well as other mitochondrial genes, for mutations that might alter CO activity and cosegregate with AD. In the present study, specific missense mutations in the mitochondrial CO1 and CO2 genes but not the CO3 gene were found to segregate at a higher frequency with AD compared with other neurodegenerative or metabolic diseases. These mutations appear together in the same mitochondrial DNA molecule and define a unique mutant mitochondrial genome. Asymptomatic offspring of AD mothers had higher levels of these mutations than offspring of AD fathers, suggesting that these mutations can be maternally inherited. Cell lines expressing these mutant mitochondrial DNA molecules exhibited a specific decrease in CO activity and increased production of reactive oxygen species. We suggest that specific point mutations in the CO1 and CO2 genes cause the CO defect in AD. A CO defect may represent a primary etiologic event, directly participating in a cascade of events that results in AD.
Figures
Figure 1
Clonal analysis of CO1 and CO2 genes from a typical AD patient showing sites of heteroplasmic mutations in mtDNA. mtDNA was prepared and cloned into appropriate vectors as described. Ten independent clones of each gene were subjected to automated sequencing. Five missense mutations and one silent mutation are represented. Black boxes indicate sites of point mutations. Open boxes indicate sites of wild-type bases. As can be seen, 50% of the clones for the CO1 gene contained a least one point mutation, whereas 30% of the clones for the CO2 gene contained at least one mutation. In most instances, these mutations appear together in the same clone suggesting a unique mutated mitochondrial genome.
Figure 2
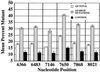
Quantitative analysis of the relative amounts of each of the six mutations identified by competitive primer extension assays. Each bar represents the group mean percentage of the mutant base relative to the wild-type base. AD total group represents the entire AD population studied. The AD high mutations group represents the mutational load at each mutant base for AD cases with levels of the mutant molecule exceeding those of any control case. The AD high mutation group included ≈20% of the AD total population. Error bars represent the SEM for each group. At each site, AD cases had significantly higher levels of the mutant base than controls as determined by independent t tests (P < 0.001).
Figure 3
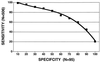
Receiver operating characteristic (ROC) curve presenting the sensitivity (percentage of AD cases with the relative proportions of the 7650 mutant base above a selected value) and specificity (percentage of non-AD cases with the relative proportions of the 7650 mutant base below a selected value). From this curve, it can be seen that 20% of all AD cases have levels of this mutant allele exceeding those of any control case (20% sensitivity, 100% specific).
Figure 4
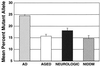
Comparison of the mutational load at the 7650 nucleotide position in AD cases, cognitively normal, age-matched controls (AGED), neurologic disease controls (NEUROLOGIC), and NIDDM patients. All disease patients and controls had significantly lower levels of this mutant allele than AD cases. There were no statistically significant differences among the three control groups.
Figure 5
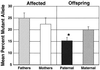
Mean percentage mutant molecules for the 7650 nucleotide position in a subset of the total sampled population for cases in which an affected parent and asymptomatic offspring pair were available. Each bar represents the group mean. Affected parents had similar levels of the 7650 mutant base, as seen on the left side of the graph. Offspring of affected mothers had higher levels of this mutant base than offspring of affected fathers, as seen on the right side of the graph. This suggests that in this subset, these mutations are transmitted through a maternal lineage.
Figure 6
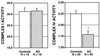
Characterization of complex I and CO activity in AD and control cybrids expressed as a rate for complex I (nmol⋅min−1⋅mg−1) and a relative rate for CO (min−1⋅mg−1). CO activity was significantly decreased in AD cybrids relative to control cybrids (t test, P < 0.001). In contrast, complex I activity, another complex encoded in part by the mitochondrial genome, was not different in a subset of AD and normal control cybrids. Each bar represents the mean group activity. Error bars are SEM. To confirm that cells were true cybrids and not hybrids, putative cybrid cells were cultured and harvested using standard cytogenetic techniques. They were then stained with the GTG (Giemsa–trypsin–Giemsa) technique, and chromosomes were counted.
Figure 7
Intracellular generation of reactive oxygen species was measured in a subset of the age-matched, cognitively normal control and AD cybrids. Each bar represents the group mean percent change from parental SH-SY5Y cells in DCF-DA fluorescence (relative mean fluorescence/cell number). AD cybrids produced significantly more reactive oxygen species than control cybrids.
Similar articles
- Mutations in mitochondrial-encoded cytochrome c oxidase subunits I, II, and III genes detected in Alzheimer's disease using single-strand conformation polymorphism.
Hamblet NS, Ragland B, Ali M, Conyers B, Castora FJ. Hamblet NS, et al. Electrophoresis. 2006 Feb;27(2):398-408. doi: 10.1002/elps.200500420. Electrophoresis. 2006. PMID: 16358358 - Mitochondrial DNA mutations in Alzheimer's disease.
Hutchin TP, Heath PR, Pearson RC, Sinclair AJ. Hutchin TP, et al. Biochem Biophys Res Commun. 1997 Dec 18;241(2):221-5. doi: 10.1006/bbrc.1997.7793. Biochem Biophys Res Commun. 1997. PMID: 9425253 - Mitochondrial genomic contribution to mitochondrial dysfunction in Alzheimer's disease.
Onyango I, Khan S, Miller B, Swerdlow R, Trimmer P, Bennett P Jr. Onyango I, et al. J Alzheimers Dis. 2006 Jul;9(2):183-93. doi: 10.3233/jad-2006-9210. J Alzheimers Dis. 2006. PMID: 16873965 Review.
Cited by
- Identification of a new missense mutation in the mtDNA of hereditary hypertrophic, but not dilated cardiomyopathic hamsters.
Minieri M, Zingarelli M, Shubeita H, Vecchini A, Binaglia L, Carotenuto F, Fantini C, Fiaccavento R, Masuelli L, Coletti A, Simonelli L, Modesti A, Di Nardo P. Minieri M, et al. Mol Cell Biochem. 2003 Oct;252(1-2):73-81. doi: 10.1023/a:1025542731335. Mol Cell Biochem. 2003. PMID: 14577578 - The pseudo-mitochondrial genome influences mistakes in heteroplasmy interpretation.
Parr RL, Maki J, Reguly B, Dakubo GD, Aguirre A, Wittock R, Robinson K, Jakupciak JP, Thayer RE. Parr RL, et al. BMC Genomics. 2006 Jul 21;7:185. doi: 10.1186/1471-2164-7-185. BMC Genomics. 2006. PMID: 16859552 Free PMC article. - Specificity of mtDNA-directed PCR-influence of NUclear MTDNA insertion (NUMT) contamination in routine samples and techniques.
Goios A, Prieto L, Amorim A, Pereira L. Goios A, et al. Int J Legal Med. 2008 Jul;122(4):341-5. doi: 10.1007/s00414-007-0191-5. Epub 2007 Sep 14. Int J Legal Med. 2008. PMID: 17874117 - The role of mitochondria in the pathogenesis of neurodegenerative diseases.
Manfredi G, Beal MF. Manfredi G, et al. Brain Pathol. 2000 Jul;10(3):462-72. doi: 10.1111/j.1750-3639.2000.tb00278.x. Brain Pathol. 2000. PMID: 10885665 Free PMC article. Review. - Ethanol withdrawal hastens the aging of cytochrome c oxidase.
Jung ME, Ju X, Metzger DB, Simpkins JW. Jung ME, et al. Neurobiol Aging. 2012 Mar;33(3):618.e21-32. doi: 10.1016/j.neurobiolaging.2011.02.002. Epub 2011 Mar 24. Neurobiol Aging. 2012. PMID: 21439684 Free PMC article.
References
- Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, et al. Nature (London) 1995;375:754–760. - PubMed
- Levy-Lahad E, Wasco W, Poorkaj P, Romano D M, Ohshima J, et al. Science. 1995;269:973–977. - PubMed
- Goate A M, Chartier M C, Mullian M, Brown J, Crawford F, et al. Nature (London) 1991;349:704–706. - PubMed
- Silverman J M, Li G, Zaccario M L, Smith C J, Schmeidler J, Mohs R C, Davis K L. Arch Gen Psychiatry. 1994;51:577–586. - PubMed
- Silverman J M, Raiford K, Edland S, Fillenbaum G, Morris J C, Clark C M, Kukull W, Heyman A. Neurology. 1994;44:1253–1259. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases