The La antigen binds 5' noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation - PubMed (original) (raw)
The La antigen binds 5' noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation
N Ali et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Translation initiation of the hepatitis C virus (HCV) RNA genome occurs through an internal ribosome entry site in a cap-independent manner. Here, we have examined the interaction between La antigen and the HCV 5' noncoding region (5'NCR). In this analysis, competitor RNAs derived from HCV 5'NCR carrying deletions and a point mutation were used to identify the site(s) of La antigen binding during UV cross-linking assay. These studies suggest that La antigen recognizes the intact HCV 5'NCR structure. Further, these interactions occurred in the context of the initiator AUG. The latter view is supported by an analysis in which mutants of the HCV 5'NCR RNA with deletion or substitution in the initiator AUG codon failed to compete for La antigen binding to the wild-type 5'NCR. The evidence for the interaction between liver cell-derived La antigen and the HCV 5'NCR is provided by immunoprecipitation of a UV cross-linked species from the S100 fraction of Huh7 cell lysates. The functional relevance of this interaction was demonstrated by the stimulation of the HCV internal ribosome entry site-mediated translation in the presence of La protein. These results suggest an important functional role of La protein in the regulation of internal initiation of translation of the HCV RNA genome.
Figures
Figure 1
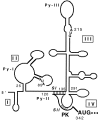
Schematic representation of computer-generated RNA folding model as proposed by Brown et al. (17) with a modification in the vicinity of initiator AUG according to Wang et al. (19). The stem I (SI) and stem II (SII) of the pseudoknot (PK) structure are shaded.
Figure 2
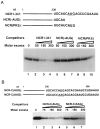
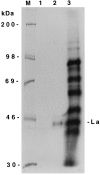
Purification of bacterially expressed human La protein and competition assay for the binding of La antigen with the full-length wild-type HCV 5′NCR. (A) SDS/PAGE and silver staining of poly(U)-Sepharose 4B affinity purified La antigen. Seventy-five nanograms of protein was loaded in lane 1. Lane M, molecular weight markers. (B) Direct UV cross-linking of the purified La preparation (shown in Fig. 2_A_, lane 1) with the wild-type 5′NCR RNA probe followed by 12% SDS/PAGE. The probe contained 4-thio-U and was labeled with [32P]CTP. The binding reactions were carried out with 15 ng of purified La (lane 2). Lane 1, probe alone. A similar UV cross-linking assay was carried out as shown in lane 2 followed by immunoprecipitation (IP) with monoclonal anti-La antibody, SW5 (lane 4), or normal serum (lane 3). The samples were fractionated on 12% SDS/PAGE and autoradiographed. (C) Effect of unlabeled competitor RNAs (homologous and deletion mutants) during the interaction of La protein with the thio-U-containing 32P-labeled NCR1–341 RNA probe. Lane 1, probe alone; lane 2, no competitor RNA; lanes 3–6, increasing amounts of unlabeled homologous RNA. The UV cross-linked samples presented from lanes 7 to 16 were fractionated on a separate SDS/polyacrylamide gel. Sample in lane 7 is same as in lane 2. Lanes 8–16 represent deletion mutants of the 5′NCR RNA used as competitors. The numbers indicate the length of nucleotide sequences of the 5′NCR. (D) Competition assay with the HCV 5′NCR mutants lacking domain III structure. UV cross-linking of the NCR1–341 RNA probe with La antigen was carried out in presence of unlabeled competitor RNA as described in the legend to Fig. 2_B_. Lane 1, no competitor RNA. Lanes 2–7, unlabeled competitor RNAs derived from 5′NCR as indicated. The T7LUC1–99 RNA used as a competitor (lanes 8, 9) represents 99 nucleotides derived from the luciferase gene. (E) Effect of a heterologous competitor RNA on the La binding to the 5′NCR RNA probe. The competitor RNAs, globin mRNA (GIBCO) (lanes 5 and 6), and homologous RNA (lanes 3 and 4), were included in the reaction mixture during UV cross-linking as described above. Lane 1, probe alone. Lane 2, no competitor RNA.
Figure 3
Effect of deletion and substitution of initiator AUG codon (iAUG) on the binding of La antigen to the 32P-labeled HCV 5′NCR. (A) Competition assay using HCV 5′NCR derived mutant RNA lacking nucleotide sequences at the translation initiation site. (Upper) Nucleotide sequences at the 3′ end of each competitor RNA. The iAUG is underlined. (Lower) Results of competition assay. The NCR(1–341) (lanes 2–4), NCR(-AUG) (lanes 5–7), and NCR(PKS) (lanes 8–10) unlabeled RNAs were used as competitors during UV cross-linking of the wild-type RNA probe (NCR1–341) with La antigen. Lane 1, no competitor RNA. (B) Effect of substitution (U to A) at the initiator AUG on La binding. (Upper) Nucleotide sequences at the 3′ end of the unlabeled RNAs used during competition assay. (Lower) The NCR-C(AUG) RNA probe was UV cross-linked with La antigen in the presence of unlabeled homologous (lanes 2–4) and the mutant NCR-C(AAG) (lanes 5–7) RNAs. Lane 1, no competitor RNA. The UV cross-linked products were fractionated by SDS/PAGE and autoradiographed.
Figure 4
Immunoprecipitation of La antigen from liver cell-derived (Huh7) S100 fraction after UV cross-linking with the 32P-labeled HCV 5′NCR RNA probe. UV cross-linked S100 lysates (lane 3) were subjected to immunoprecipitation with monoclonal anti-La antibody (SW5) (lane 2) or normal rabbit serum (lane 1). The samples were fractionated by SDS/PAGE.
Figure 5
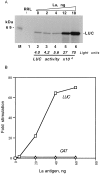
La antigen stimulates translation of the HCV IRES-controlled luciferase RNA in RRL. (A) Translation of monocistronic RNA (T7C1–341) with increasing amounts of purified La antigen (lanes 3–6) or without added La antigen (lane 2). The translation was carried out with (lanes 2–6) or without (lane 1) T7C1–341 RNA (0.2 μg) in RRL. The translation products were radiolabeled with [35S]methionine. The final volume (20 μl) of the reaction mixtures was adjusted with buffer D. Five microliters of translation mixtures was fractionated by SDS/PAGE. The gels were treated with Fluoro-Hance (Research Products International) before autoradiography. Luciferase activity was measured with a 2 μl aliquot of the translation mixture according to de Wet et al. (38). (B) Stimulation of HCV IRES activity in the presence of recombinant La antigen. The dicistronic RNA template (T7DC1–341) used for translation as described in A contained HCV 5′NCR between upstream chloramphenicol acetyltransferase (CAT) and downstream luciferase (LUC) cistrons. Increasing amounts of purified La antigen were added to the translation mixture. The translation mixtures were fractionated by SDS/ PAGE. The band intensities of the LUC and CAT expression were measured by densitometry using PhosphorImager. The fold-stimulation of translation was calculated using these data and plotted against the recombinant La antigen added to the translation mixtures.
Similar articles
- Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation.
Ali N, Pruijn GJ, Kenan DJ, Keene JD, Siddiqui A. Ali N, et al. J Biol Chem. 2000 Sep 8;275(36):27531-40. doi: 10.1074/jbc.M001487200. J Biol Chem. 2000. PMID: 10856291 - Complete 5' noncoding region is necessary for the efficient internal initiation of hepatitis C virus RNA.
Fukushi S, Katayama K, Kurihara C, Ishiyama N, Hoshino FB, Ando T, Oya A. Fukushi S, et al. Biochem Biophys Res Commun. 1994 Mar 15;199(2):425-32. doi: 10.1006/bbrc.1994.1246. Biochem Biophys Res Commun. 1994. PMID: 8135783 - Translation of hepatitis C virus genome.
Ali N, Wang C, Siddiqui A. Ali N, et al. Princess Takamatsu Symp. 1995;25:99-110. Princess Takamatsu Symp. 1995. PMID: 8875614 Review. - Mechanism of translation initiation on hepatitis C virus RNA.
Nomoto A, Tsukiyama-Kohara K, Kohara M. Nomoto A, et al. Princess Takamatsu Symp. 1995;25:111-9. Princess Takamatsu Symp. 1995. PMID: 8875615 Review.
Cited by
- Inhibition of the protein kinase PKR by the internal ribosome entry site of hepatitis C virus genomic RNA.
Vyas J, Elia A, Clemens MJ. Vyas J, et al. RNA. 2003 Jul;9(7):858-70. doi: 10.1261/rna.5330503. RNA. 2003. PMID: 12810919 Free PMC article. - A cell-permeable peptide inhibits hepatitis C virus replication by sequestering IRES transacting factors.
Fontanes V, Raychaudhuri S, Dasgupta A. Fontanes V, et al. Virology. 2009 Nov 10;394(1):82-90. doi: 10.1016/j.virol.2009.08.012. Epub 2009 Sep 8. Virology. 2009. PMID: 19740508 Free PMC article. - Novel fluorescence-based screen to identify small synthetic internal ribosome entry site elements.
Venkatesan A, Dasgupta A. Venkatesan A, et al. Mol Cell Biol. 2001 Apr;21(8):2826-37. doi: 10.1128/MCB.21.8.2826-2837.2001. Mol Cell Biol. 2001. PMID: 11283261 Free PMC article. - Sam68 Promotes Hepatitis C Virus Replication by Interaction with Stem-Loop 2 of Viral 5' Untranslated Region.
Qin Y, Xun Z, Guo Y, Chen S, Zhu H. Qin Y, et al. J Virol. 2019 Jun 28;93(14):e00693-19. doi: 10.1128/JVI.00693-19. Print 2019 Jul 15. J Virol. 2019. PMID: 31068419 Free PMC article. - The levels of the RoRNP-associated Y RNA are dependent upon the presence of ROP-1, the Caenorhabditis elegans Ro60 protein.
Labbé JC, Hekimi S, Rokeach LA. Labbé JC, et al. Genetics. 1999 Jan;151(1):143-50. doi: 10.1093/genetics/151.1.143. Genetics. 1999. PMID: 9872955 Free PMC article.
References
- Tsukuma H, Hiyama T, Tanaka S, Nakao M, Yabuuchi T, Kitamura T, Nakanishi K, Fujimoto I, Inoue A, Yamazaki H, Kawashima T. N Engl J Med. 1993;328:1797–1801. - PubMed
- Houghton M, Weiner A, Han J, Kuo G, Choo Q-L. Hepatology. 1991;14:381–388. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous