The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae - PubMed (original) (raw)
The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae
J de la Cruz et al. Proc Natl Acad Sci U S A. 1997.
Abstract
The translation initiation factor eIF4E mediates the binding of the small ribosomal subunit to the cap structure at the 5' end of the mRNA. In Saccharomyces cerevisiae, the cap-binding protein eIF4E is mainly associated with eIF4G, forming the cap-binding complex eIF4F. Other proteins are detected upon purification of the complex on cap-affinity columns. Among them is p20, a protein of unknown function encoded by the CAF20 gene. Here, we show a negative regulatory role for the p20 protein in translation initiation. Deletion of CAF20 partially suppresses mutations in translation initiation factors. Overexpression of the p20 protein results in a synthetic enhancement of translation mutation phenotypes. Similar effects are observed for mutations in the DED1 gene, which we have isolated as a multicopy suppressor of a temperature-sensitive eIF4E mutation. The DED1 gene encodes a putative RNA helicase of the DEAD-box family. The analyses of its suppressor activity, of polysome profiles of ded1 mutant strains, and of synthetic lethal interactions with different translation mutants indicate that the Ded1 protein has a role in translation initiation in S. cerevisiae.
Figures
Figure 1
Suppression of the Δstm1 phenotype by Δcaf20. CDK36–1A (Δcaf20) and RCB1–1C (Δstm1) were crossed and subsequently sporulated. A representative tetratype tetrad is shown on rich medium (yeast peptone dextrose) at 30°C and 18°C. The plates were incubated for 3 and 6 days, respectively.
Figure 2
Inhibition of Δstm1 growth by p20 overexpression. The RCB1–1C (Δstm1) strain was transformed with either the pGAL-p20 (_CEN_-URA3) or YEPL1-p20 (2μ-URA3) plasmids that contain the CAF20 gene under a _CYC1_-GAL1 promoter or as a control with pGAL or YEPL1. Transformants were grown for 4 days on SD-Ura or on SGal-Ura at 30°C.
Figure 3
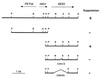
The PET56-DED1 region of chromosome XV. The original fragment carrying the suppressor activity and four relevant subclones are shown. B, _Bam_HI; H, _Hin_dIII; P, _Pst_I; S, _Sal_I; X, _Xba_I. The _Sal_I site belongs to the polylinker of YEplac181.
Figure 4
Suppression of the cdc33–42 mutation by DED1 and DBP1. CDK35–4A was transformed with either YEplac181, YEplac181-CDC33, YEplac181-DED1, or YEplac181-DBP1. Transformants were grown on SD-Leu at 30°C or 35°C for 3 and 6 days, respectively.
Figure 5
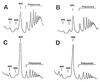
Polysome analysis of ded1 and dbp1 mutants. Cells were grown in yeast peptone dextrose at 30°C and harvested at an OD600 of 0.8. The peaks of free 40S and 60S ribosomal subunits, 80S, and polysomes are indicated. (A) DBY747, wild-type strain. (B) CDK112–1A, Δdpb1 strain. (C) DJY105, ded1/spp81–3 mutant. (D) DJY106, ded1/spp81–2 mutant.
Similar articles
- A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E.
Altmann M, Schmitz N, Berset C, Trachsel H. Altmann M, et al. EMBO J. 1997 Mar 3;16(5):1114-21. doi: 10.1093/emboj/16.5.1114. EMBO J. 1997. PMID: 9118949 Free PMC article. - Distinct interactions of eIF4A and eIF4E with RNA helicase Ded1 stimulate translation in vivo.
Gulay S, Gupta N, Lorsch JR, Hinnebusch AG. Gulay S, et al. Elife. 2020 May 29;9:e58243. doi: 10.7554/eLife.58243. Elife. 2020. PMID: 32469309 Free PMC article. - Modulation of eukaryotic mRNA stability via the cap-binding translation complex eIF4F.
Ramirez CV, Vilela C, Berthelot K, McCarthy JE. Ramirez CV, et al. J Mol Biol. 2002 May 10;318(4):951-62. doi: 10.1016/S0022-2836(02)00162-6. J Mol Biol. 2002. PMID: 12054793 - Eukaryotic translation initiation: there are (at least) two sides to every story.
Sachs AB, Varani G. Sachs AB, et al. Nat Struct Biol. 2000 May;7(5):356-61. doi: 10.1038/75120. Nat Struct Biol. 2000. PMID: 10802729 Review. - eIF4E and Interactors from Unicellular Eukaryotes.
Ross-Kaschitza D, Altmann M. Ross-Kaschitza D, et al. Int J Mol Sci. 2020 Mar 21;21(6):2170. doi: 10.3390/ijms21062170. Int J Mol Sci. 2020. PMID: 32245232 Free PMC article. Review.
Cited by
- DEAD box RNA helicase functions in cancer.
Fuller-Pace FV. Fuller-Pace FV. RNA Biol. 2013 Jan;10(1):121-32. doi: 10.4161/rna.23312. Epub 2013 Jan 1. RNA Biol. 2013. PMID: 23353573 Free PMC article. Review. - Coupling between the DEAD-box RNA helicases Ded1p and eIF4A.
Gao Z, Putnam AA, Bowers HA, Guenther UP, Ye X, Kindsfather A, Hilliker AK, Jankowsky E. Gao Z, et al. Elife. 2016 Aug 5;5:e16408. doi: 10.7554/eLife.16408. Elife. 2016. PMID: 27494274 Free PMC article. - Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA.
Tarun SZ Jr, Sachs AB. Tarun SZ Jr, et al. Mol Cell Biol. 1997 Dec;17(12):6876-86. doi: 10.1128/MCB.17.12.6876. Mol Cell Biol. 1997. PMID: 9372919 Free PMC article. - The RNA Helicase Ded1 from Yeast Is Associated with the Signal Recognition Particle and Is Regulated by SRP21.
Yeter-Alat H, Belgareh-Touzé N, Le Saux A, Huvelle E, Mokdadi M, Banroques J, Tanner NK. Yeter-Alat H, et al. Molecules. 2024 Jun 20;29(12):2944. doi: 10.3390/molecules29122944. Molecules. 2024. PMID: 38931009 Free PMC article. - Mechanism and Regulation of Protein Synthesis in Saccharomyces cerevisiae.
Dever TE, Kinzy TG, Pavitt GD. Dever TE, et al. Genetics. 2016 May;203(1):65-107. doi: 10.1534/genetics.115.186221. Genetics. 2016. PMID: 27183566 Free PMC article. Review.
References
- Merrick W C, Hershey J W B. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 31–69.
- Iizuka N, Chen C, Yang Q, Johannes G, Sarnow P. Curr Top Microbiol Immunol. 1995;203:155–177. - PubMed
- Jaramillo M, Browning K, Dever T E, Blum S, Trachsel H, Merrick W C, Ravel J M, Sonenberg N. Biochim Biophys Acta. 1990;1050:134–139. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous