Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway - PubMed (original) (raw)
Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway
J Kim et al. J Cell Biol. 1997.
Abstract
Aminopeptidase I (API) is transported into the yeast vacuole by the cytoplasm to vacuole targeting (Cvt) pathway. Genetic evidence suggests that autophagy, a major degradative pathway in eukaryotes, and the Cvt pathway share largely the same cellular machinery. To understand the mechanism of the Cvt import process, we examined the native state of API. Dodecameric assembly of precursor API in the cytoplasm and membrane binding were rapid events, whereas subsequent vacuolar import appeared to be rate limiting. A unique temperature-sensitive API-targeting mutant allowed us to kinetically monitor its oligomeric state during translocation. Our findings indicate that API is maintained as a dodecamer throughout its import and will be useful to study the posttranslational movement of folded proteins across biological membranes.
Figures
Figure 1
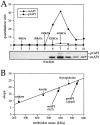
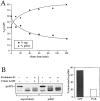
Molecular mass determination of native precursor and mature API under steady-state conditions. (A) Glycerol gradient analysis of precursor and mature API under steady-state conditions. Wild-type (SEY6210) cells were grown to midlog phase, lysed with glass beads, and the resulting cell extracts were separated on 20–50% glycerol gradients. Collected fractions were subjected to Western blotting with antiserum to API. The reaction of a chemifluorescent substrate (ECF) with an alkaline phosphatase– conjugated secondary antibody allowed for the quantitation of the Western blots by a chemifluorescence scanner (Molecular Dynamics) as shown in the graph corresponding to the Western blot signals. Molecular mass standards indicated are hen egg albumin (45 kD), aldolase (158 kD), catalase (240 kD), ferritin (450 kD), and thyroglobulin (669 kD). Both mature (mAPI) and precursor API (prAPI) peaks appeared in fraction 7, cofractionating with the thyroglobulin molecular mass standard. (B) Hedrick-Smith calculation of the molecular masses of precursor and mature API. Relative mobilities were measured for protein standards resolved on native gels of 4.0 to 5.5% acrylamide and the negative slopes were determined by plotting the relative mobilities as a function of gel percentage. A standard curve was then generated by plotting the negative slopes of the protein standards as a function of molecular mass. Extracts from wild-type and _pep4_Δ strains were run on the same gels, and their molecular masses were calculated with reference to the standard curve. Molecular masses of mAPI and precursor API were determined to be 592 and 752 kD, respectively.
Figure 2
Oligomerization kinetics of precursor API. Wild-type cells were pulse labeled for 2 min at 30°C followed by nonradioactive chase reactions. Aliquots were removed at the indicated chase times and lysed with glass beads, and the resulting cell extracts were separated on 20–50% glycerol gradients. Fractions were collected, immunoprecipitated with antiserum to API, and resolved by SDS-PAGE. Molecular mass standards corresponding to the fractions: 45 kD, fraction 2; 158 kD, fraction 4; 240 kD, fraction 5; 450 kD, fraction 6; and 669 kD, fraction 7. Quantitation of the radioactive signals was performed using a phosphorimager (model Storm; Molecular Dynamics).
Figure 3
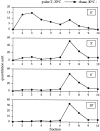
Oligomeric precursor API assembly occurs in the cytoplasm before membrane binding. Spheroplasts were labeled for 5 min at 30°C followed by a 3-min nonradioactive chase reaction. The samples were subjected to differential osmotic lysis and separated into a supernatant fraction and a pellet fraction containing intact vacuoles. An aliquot of the supernatant and pellet fractions was removed and immunoprecipitated with the indicated antisera, and the remainder was separated on 20–50% glycerol gradients. (A) Quantitation of the pellet fraction immunoprecipitated with vacuolar markers PrA and ALP, and the cytosolic marker PGK. The recovery of marker protein in the pellet was quantified using a Storm phosphorimager. The percent recovery was calculated as the ratio of the protein in the pellet fraction to the protein in the pellet and supernatant fractions. The supernatant (sup) fraction (B) and the pellet fraction (C) were separated on a glycerol gradient and immunoprecipitated with antiserum to API. Quantitation of the radioactive signals, represented by the graphs, indicates that the supernatant fraction contains both the precursor monomer and oligomer, while only the precursor API oligomer is bound to the pellet fraction.
Figure 4
API-targeting mutants are not defective in API oligomerization. Strain DYY101 (ape1_Δ) harboring a single copy plasmid encoding the propeptide deletions was grown to midlog phase, lysed with glass beads, and separated on a 20–50% glycerol gradient. The Δ_9–11 API deletion mutant shown here is a representative example of the analysis of the cvt (1 to 17) and propeptide deletion mutants (Δ_3–5_, Δ_6–8_, Δ_9–11_, Δ_12–14_, Δ_15– 17_, Δ_18–20_, Δ_25–27_, Δ_28–30_, Δ_31–33_, Δ_34–36_, Δ_37–39_, Δ_40–42_, and Δ_2–45_). Collected fractions were subjected to Western blotting with antiserum to API and the quantitation of the Western blots (shown in the graph) was performed as in Fig. 1.
Figure 5
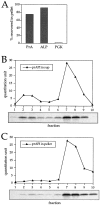
The membrane accumulation phenotype of the K12R API ts mutant is thermally reversible. (A) K12R API accumulates in the pellet fraction at nonpermissive temperature. Spheroplasts were labeled for 10 min at 38°C followed by nonradioactive chase. Aliquots were removed at the indicated chase times and separated into supernatant (sup) and pellet fractions. Samples were immunoprecipitated with antiserum to API, and the radiolabeled signals were quantitated. The percent radiolabeled precursor at a given chase point represents the ratio of precursor from each supernatant or pellet fraction to the total API combined in both fractions. (B) Protease accessibility of K12R API in the supernatant and pellet fractions. Spheroplasts were labeled for 5 min, chased for 30 min at 38°C, and separated into a supernatant and pellet fraction after differential osmotic lysis. The supernatant and pellet fractions were subjected to proteinase K and Triton X-100 as indicated and immunoprecipitated with antiserum to API (left). An aliquot of the recovered pellet fraction was also immunoprecipitated with antisera to the vacuolar marker CPY and the cytosolic marker PGK before protease treatment (right). The percent of marker proteins recovered in the pellet fraction was calculated as described for Fig. 3. (C) Accessibility of K12R API in the pellet fraction to cross-linking with Sulfo-NHS-biotin. Labeled spheroplasts were fractionated exactly as in B. The pellet fraction was cross-linked with Sulfo-NHS-biotin (Biotin-X) in the presence or absence of Triton X-100. API and CPY were recovered by immunoprecipitation followed by precipitation with avidin agarose beads. (D) Thermal reversibility of the K12R membrane-accumulation phenotype. The K12R API mutant was pulse-labeled for 10 min, chased for 30 min at 38°C, and then shifted to 30°C (top). Samples were removed at the indicated times during the shift period at 30°C and lysed with glass beads. The resulting cell extracts were immunoprecipitated with antiserum to API and resolved by SDS-PAGE. The double-headed arrow in the top panel marks the 20–40-min window of time when mature API increases from 10 to 56% during the 30°C shift. The K12R API strain was also pulse labeled, chased, and incubated all at 38°C (middle) or 30°C (bottom) in this experiment.
Figure 5
The membrane accumulation phenotype of the K12R API ts mutant is thermally reversible. (A) K12R API accumulates in the pellet fraction at nonpermissive temperature. Spheroplasts were labeled for 10 min at 38°C followed by nonradioactive chase. Aliquots were removed at the indicated chase times and separated into supernatant (sup) and pellet fractions. Samples were immunoprecipitated with antiserum to API, and the radiolabeled signals were quantitated. The percent radiolabeled precursor at a given chase point represents the ratio of precursor from each supernatant or pellet fraction to the total API combined in both fractions. (B) Protease accessibility of K12R API in the supernatant and pellet fractions. Spheroplasts were labeled for 5 min, chased for 30 min at 38°C, and separated into a supernatant and pellet fraction after differential osmotic lysis. The supernatant and pellet fractions were subjected to proteinase K and Triton X-100 as indicated and immunoprecipitated with antiserum to API (left). An aliquot of the recovered pellet fraction was also immunoprecipitated with antisera to the vacuolar marker CPY and the cytosolic marker PGK before protease treatment (right). The percent of marker proteins recovered in the pellet fraction was calculated as described for Fig. 3. (C) Accessibility of K12R API in the pellet fraction to cross-linking with Sulfo-NHS-biotin. Labeled spheroplasts were fractionated exactly as in B. The pellet fraction was cross-linked with Sulfo-NHS-biotin (Biotin-X) in the presence or absence of Triton X-100. API and CPY were recovered by immunoprecipitation followed by precipitation with avidin agarose beads. (D) Thermal reversibility of the K12R membrane-accumulation phenotype. The K12R API mutant was pulse-labeled for 10 min, chased for 30 min at 38°C, and then shifted to 30°C (top). Samples were removed at the indicated times during the shift period at 30°C and lysed with glass beads. The resulting cell extracts were immunoprecipitated with antiserum to API and resolved by SDS-PAGE. The double-headed arrow in the top panel marks the 20–40-min window of time when mature API increases from 10 to 56% during the 30°C shift. The K12R API strain was also pulse labeled, chased, and incubated all at 38°C (middle) or 30°C (bottom) in this experiment.
Figure 6
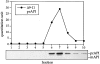
Membrane-associated API is imported into the vacuole as an oligomer. The K12R API mutant was pulse labeled for 10 min and chased for 30 min at 38°C and then shifted to 30°C. Samples were removed at the indicated times during the 20–40-min window of the 30°C shift period and lysed with glass beads, and the resulting extract was separated by 20–50% glycerol gradients. Fractions were collected, immunoprecipitated with antiserum to API, and resolved by SDS-PAGE. Both mature (mAPI) and precursor API (prAPI) peaks appeared in fraction 7, cofractionating with the thyroglobulin molecular mass standard (669 kD).
Figure 7
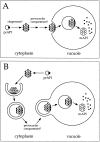
Models for API import via the Cvt pathway. Precursor API is synthesized and assembled into dodecamers in the cytoplasm, followed by membrane binding. During the rate-limiting step of the pathway, both models A and B propose a vesicle-mediated mechanism of API entry into the vacuole followed by the breakdown of the API-containing vesicles and cleavage of the API propeptide in a PrB-dependent manner to yield the mature hydrolase. The role of molecular chaperones and the location of the initial binding of precursor API remain to be resolved. (A) Oligomeric precursor API directly binds to the vacuolar or prevacuolar membrane before the vesicle-mediated entry into the vacuole. (B) Genetic analyses have revealed a large overlap between the autophagy and Cvt pathways. In this model, API is delivered to the vacuole via double-membrane autophagic vesicles. Upon reaching the vacuole, these vesicles fuse to the vacuolar membrane, releasing a single-membrane vesicle (autophagic body) containing precursor API. This is followed by the breakdown of the vesicles and subsequent processing of the hydrolase to the mature enzyme.
Similar articles
- Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome.
Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Baba M, et al. J Cell Biol. 1997 Dec 29;139(7):1687-95. doi: 10.1083/jcb.139.7.1687. J Cell Biol. 1997. PMID: 9412464 Free PMC article. - Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting, and role of the vacuole in degradation.
Teter SA, Klionsky DJ. Teter SA, et al. Semin Cell Dev Biol. 2000 Jun;11(3):173-9. doi: 10.1006/scdb.2000.0163. Semin Cell Dev Biol. 2000. PMID: 10906274 Review. - Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I.
Oda MN, Scott SV, Hefner-Gravink A, Caffarelli AD, Klionsky DJ. Oda MN, et al. J Cell Biol. 1996 Mar;132(6):999-1010. doi: 10.1083/jcb.132.6.999. J Cell Biol. 1996. PMID: 8601598 Free PMC article. - Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p.
Abeliovich H, Darsow T, Emr SD. Abeliovich H, et al. EMBO J. 1999 Nov 1;18(21):6005-16. doi: 10.1093/emboj/18.21.6005. EMBO J. 1999. PMID: 10545112 Free PMC article. - Molecular machinery required for autophagy and the cytoplasm to vacuole targeting (Cvt) pathway in S. cerevisiae.
Khalfan WA, Klionsky DJ. Khalfan WA, et al. Curr Opin Cell Biol. 2002 Aug;14(4):468-75. doi: 10.1016/s0955-0674(02)00343-5. Curr Opin Cell Biol. 2002. PMID: 12383798 Review.
Cited by
- Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion.
Reggiori F, Wang CW, Stromhaug PE, Shintani T, Klionsky DJ. Reggiori F, et al. J Biol Chem. 2003 Feb 14;278(7):5009-20. doi: 10.1074/jbc.M210436200. Epub 2002 Nov 20. J Biol Chem. 2003. PMID: 12446664 Free PMC article. - An overview of the molecular mechanism of autophagy.
Yang Z, Klionsky DJ. Yang Z, et al. Curr Top Microbiol Immunol. 2009;335:1-32. doi: 10.1007/978-3-642-00302-8_1. Curr Top Microbiol Immunol. 2009. PMID: 19802558 Free PMC article. Review. - A role for Atg8-PE deconjugation in autophagosome biogenesis.
Nair U, Yen WL, Mari M, Cao Y, Xie Z, Baba M, Reggiori F, Klionsky DJ. Nair U, et al. Autophagy. 2012 May 1;8(5):780-93. doi: 10.4161/auto.19385. Epub 2012 May 1. Autophagy. 2012. PMID: 22622160 Free PMC article. - The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy.
Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, Hughson FM, Baba M, Klionsky DJ. Yen WL, et al. J Cell Biol. 2010 Jan 11;188(1):101-14. doi: 10.1083/jcb.200904075. J Cell Biol. 2010. PMID: 20065092 Free PMC article. - Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex.
Kim J, Huang WP, Klionsky DJ. Kim J, et al. J Cell Biol. 2001 Jan 8;152(1):51-64. doi: 10.1083/jcb.152.1.51. J Cell Biol. 2001. PMID: 11149920 Free PMC article.
References
- Davis LI. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896. - PubMed
- Dunn WA., Jr Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–143. - PubMed
- Egner R, Thumm M, Straub M, Simeon A, Schuller HJ, Wolf DH. Tracing intracellular proteolytic pathways. Proteolysis of fatty acid synthase and other cytoplasmic proteins in the yeast Saccharomyces cerevisiae. . J Biol Chem. 1993;268:27269–27276. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases