SOS factors involved in translesion synthesis - PubMed (original) (raw)
SOS factors involved in translesion synthesis
R L Napolitano et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Mutations are permanent DNA sequence changes that can be induced when replication occurs on a damaged DNA template. In Escherichia coli, the process of translesion synthesis past a lesion that hinders replication requires the induction of SOS-controlled gene products, among which are those of the umuDC operon. To study translesion synthesis in vivo, we have constructed single-stranded vectors containing single 2-acetylaminofluorene adducts located within -1 and -2 frameshift mutation hot spots formed by short repetitive sequences. These adducts strongly hinder DNA replication as only 2-5% of the molecules give rise to progeny under non-SOS-induced conditions. Induction of the SOS response lead to a 10-fold increase in survival. Adducts present within repetitive sequences trigger the formation of misaligned primer/template replication intermediates which, upon elongation, will result in the fixation of frameshift errors (mutagenic translesion synthesis). Surprisingly we find that elongation from the nonslipped intermediate depends upon functional umuDC+ gene products, whereas elongation from the slipped intermediate is umuDC+ independent but requires another, as yet biochemically uncharacterized, SOS function. These data are discussed in terms of the different steps involved during translesion synthesis through a replication-blocking lesion.
Figures
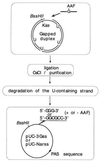
Figure 1
Strategy for the construction of single-stranded DNA. Gapped-duplex intermediates were produced as previously described (20, 24). The gapped strand of the duplex was provided by a plasmid that was grown in a wild-type strain, whereas the complementary strand contained several uracil residues as the corresponding plasmid was propagated in a dut, ung strain (CJ 236) (25). Single AAF-adducted oligonucleotides were ligated into the gap, and the covalently closed circular plasmid was purified from CsCl equilibrium sedimentation. The uracil-containing strand was then selectively degraded using an enzymatic mixture containing uracil DNA glycosylase, and the 3′ → 5′ exonuclease of exonucleaseIII and T7 DNA polymerase (25). Constructions containing the G-AAF adduct (G*) within the sequence 5′-GGG*- or 5′-GGCG*CC- were made along with the corresponding control vectors (no adduct).
Figure 2
Structure of replication intermediates and involvement of SOS factors. (A) AAF-induced frameshift mutagenesis. When AAF adducts are situated in repetitive sequence contexts associated with both −1 and −2 frameshifts, the TLS intermediates can assume two forms: a nonslipped intermediate or a slipped intermediate. Elongation from the nonslipped intermediate, i.e., error-free elongation, proceeds from a lesion terminus at which the terminal nucleotide of the nascent strand is situated across from the AAF lesion (circled). Elongation from the lesion terminus appears to be stimulated by UmuDC. In contrast, frameshift mutations result from the elongation of the postlesion terminus of the slipped intermediate, at which the terminal nucleotide of the nascent strand is correctly paired with residues situated 5′ to the adduct on the template strand (shown in the square). G* is the AAF adducted guanine residue. (B) UV-induced base substitution mutagenesis. Both error-free and mutagenic TLS across a photoproduct involve elongation from a lesion terminus because, in the intermediate, the terminal nucleotide in the nascent strand is situated across from the lesion (circled). Both elongation events are strongly stimulated by the presence of UmuDC.
Similar articles
- Inactivation of DNA proofreading obviates the need for SOS induction in frameshift mutagenesis.
Fuchs RP, Napolitano RL. Fuchs RP, et al. Proc Natl Acad Sci U S A. 1998 Oct 27;95(22):13114-9. doi: 10.1073/pnas.95.22.13114. Proc Natl Acad Sci U S A. 1998. PMID: 9789050 Free PMC article. - Sequence-dependent modulation of frameshift mutagenesis at NarI-derived mutation hot spots.
Broschard TH, Koffel-Schwartz N, Fuchs RP. Broschard TH, et al. J Mol Biol. 1999 Apr 23;288(1):191-9. doi: 10.1006/jmbi.1999.2667. J Mol Biol. 1999. PMID: 10329136 - All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis.
Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. Napolitano R, et al. EMBO J. 2000 Nov 15;19(22):6259-65. doi: 10.1093/emboj/19.22.6259. EMBO J. 2000. PMID: 11080171 Free PMC article. - Translesion synthesis in Escherichia coli: lessons from the NarI mutation hot spot.
Fuchs RP, Fujii S. Fuchs RP, et al. DNA Repair (Amst). 2007 Jul 1;6(7):1032-41. doi: 10.1016/j.dnarep.2007.02.021. Epub 2007 Apr 2. DNA Repair (Amst). 2007. PMID: 17403618 Review. - Mutagenesis and more: umuDC and the Escherichia coli SOS response.
Smith BT, Walker GC. Smith BT, et al. Genetics. 1998 Apr;148(4):1599-610. doi: 10.1093/genetics/148.4.1599. Genetics. 1998. PMID: 9560379 Free PMC article. Review.
Cited by
- Adaptive Gene Loss? Tracing Back the Pseudogenization of the Rabbit CCL8 Chemokine.
van der Loo W, Magalhaes MJ, de Matos AL, Abrantes J, Yamada F, Esteves PJ. van der Loo W, et al. J Mol Evol. 2016 Aug;83(1-2):12-25. doi: 10.1007/s00239-016-9747-7. Epub 2016 Jun 15. J Mol Evol. 2016. PMID: 27306379 - Repair of chromosomal abasic sites in vivo involves at least three different repair pathways.
Otterlei M, Kavli B, Standal R, Skjelbred C, Bharati S, Krokan HE. Otterlei M, et al. EMBO J. 2000 Oct 16;19(20):5542-51. doi: 10.1093/emboj/19.20.5542. EMBO J. 2000. PMID: 11032821 Free PMC article. - Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD'2C mutagenic complex and RecA protein.
Tang M, Bruck I, Eritja R, Turner J, Frank EG, Woodgate R, O'Donnell M, Goodman MF. Tang M, et al. Proc Natl Acad Sci U S A. 1998 Aug 18;95(17):9755-60. doi: 10.1073/pnas.95.17.9755. Proc Natl Acad Sci U S A. 1998. PMID: 9707548 Free PMC article. - Development of a versatile high-throughput mutagenesis assay with multiplexed short-read NGS using DNA-barcoded supF shuttle vector library amplified in E. coli.
Kawai H, Iwata R, Ebi S, Sugihara R, Masuda S, Fujiwara C, Kimura S, Kamiya H. Kawai H, et al. Elife. 2022 Oct 10;11:e83780. doi: 10.7554/eLife.83780. Elife. 2022. PMID: 36214452 Free PMC article. - Antagonism of ultraviolet-light mutagenesis by the methyl-directed mismatch-repair system of Escherichia coli.
Liu H, Hewitt SR, Hays JB. Liu H, et al. Genetics. 2000 Feb;154(2):503-12. doi: 10.1093/genetics/154.2.503. Genetics. 2000. PMID: 10655206 Free PMC article.
References
- Livneh Z, Cohen-Fix O, Skaliter R, Elizur T. Crit Rev Biochem Mol Biol. 1993;28:465–513. - PubMed
- Higgins N P, Kato K, Strauss B. J Mol Biol. 1976;101:417–425. - PubMed
- Rupp W D, Wilde C E, III, Reno D L, Howard-Flanders P. J Mol Biol. 1971;61:25–44. - PubMed
- Belguise-Valladier P, Maki H, Sekiguchi M, Fuchs R P P. J Mol Biol. 1994;236:151–164. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous