Positional cloning of the mouse circadian clock gene - PubMed (original) (raw)
. 1997 May 16;89(4):641-53.
doi: 10.1016/s0092-8674(00)80245-7.
Y Zhao, A M Sangoram, L D Wilsbacher, M Tanaka, M P Antoch, T D Steeves, M H Vitaterna, J M Kornhauser, P L Lowrey, F W Turek, J S Takahashi
Affiliations
- PMID: 9160755
- PMCID: PMC3815553
- DOI: 10.1016/s0092-8674(00)80245-7
Positional cloning of the mouse circadian clock gene
D P King et al. Cell. 1997.
Abstract
We used positional cloning to identify the circadian Clock gene in mice. Clock is a large transcription unit with 24 exons spanning approximately 100,000 bp of DNA from which transcript classes of 7.5 and approximately 10 kb arise. Clock encodes a novel member of the bHLH-PAS family of transcription factors. In the Clock mutant allele, an A-->T nucleotide transversion in a splice donor site causes exon skipping and deletion of 51 amino acids in the CLOCK protein. Clock is a unique gene with known circadian function and with features predicting DNA binding, protein dimerization, and activation domains. CLOCK represents the second example of a PAS domain-containing clock protein (besides Drosophila PERIOD), which suggests that this motif may define an evolutionarily conserved feature of the circadian clock mechanism.
Figures
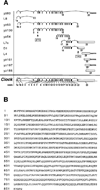
Figure 1. Genetic, Physical, and Transcription Unit Mapping of the Clock Locus
(A) Genetic map of mouse chromosome 5, showing the location of Clock on the midportion of this chromosome, 0.7 cM distal of Kit, flanked by D5Mit307 and D5Mit112. (B) Physical and high resolution genetic mapping of the Clock locus. The SSLPs and STSs in the _D5Mit307_–D5Mit112 interval are shown on the upper bar. The maximum nonrecombinant interval is defined by the SSLPs D5Mit307 and D5Nwu2. The animals with recombinations between these SSLPs and Clock are noted. Also shown are restriction sites for NotI (depicted with an [N]), which identify two CpG islands associated with the 5′ ends of Clock and pFT27. YAC and BAC contigs of the Clock region are shown below. Only four clones from the 32 clone YAC contig are shown. YACs 18 and 12 are chimeric clones and appear to be identical to YACs B16.S5.RE.C12 and B14.S4.RH.C8 in Brunkow et al. (1995); the part of each clone not mapping to this region of chromosome 5 is represented by a dashed line. The YACs were isolated from a library constructed by Kusumi et al. (1993), except for clone 55, which came from a library constructed by Larin et al. (1991). The BAC clones were isolated from a library constructed in Dr. Melvin Simon’s laboratory. (C) Candidate genes mapping to the nonrecombinant interval. Shown are their relative locations and transcriptional orientations.
Figure 2. RNA Expression of the PAS Domain Candidate Gene in
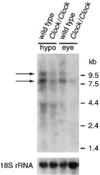
Clock/Clock Mice Differs from the Wild Type Northern blot analysis of RNA extracted (at ZT6) from eye and hypothalamus of (BALB/cJ x C57BL/6J)F2 and F3 wild-type and Clock/ Clock mice, using the yz50 cDNA clone as probe, reveals a reduction in the level of mRNA expression of both the ~8 kb and ~11 kb transcripts. The positions to which standards migrated and their sizes are shown on the right. The bottom panel shows the same filter hybridized with a 32P-labeled oligonucleotide of 18S RNA for normalization.
Figure 3. cDNA Clones Identifying the Clock Gene and the Predicted Amino Acid Sequence of the CLOCK Protein
(A) cDNA clones isolated in the cDNA selection and direct library screening efforts. Clones beginning with yz are derived from cDNA selection, and those beginning with L (LIGHT) or D (DARK) are clones isolated from the direct library screen. Their exon content was determined by alignment to the genomic sequence that was assembled from the M13 shotgun sequencing subclones of BACs 52 and 54. Exon and intron sizes are depicted schematically. Exons 1a–22 vary in size from 71 base pairs (exon 1a) to 295 base pairs (exon 22). Intron sizes vary from 107 base pairs (between exons 10 and 11) to >22 kilobase pairs (between exons 2 and 3). yz80 appears to contain a unique splice variant. L7c appears to be a partial clone of the Clock mutant splice variant transcript. Poly(A) tails of clones yz80, L4, yz161, and yz197 are indicated schematically. Exons containing open reading frames are closed, and exons containing 5′ and 3′ un-translated regions are open. Clones L8 and yz130 were reverse transcribed from intronic poly(A) sequences. The intronic sequences of these clones are shaded. The 2565-base ORF begins at base 44 of exon 4 and ends at base 177 of exon 23. Note the long 3′ un-translated region, which extends for >6 kb. (B) When translated, the 2565-base ORF predicts a protein of 855 amino acids, dominated by glutamine (112) and serine (100) residues. This protein has an expected molecular mass of 96.4 kDa and a pI of 6.52.
Figure 4. CLOCK Encodes a Novel Member of the bHLH–PAS Family of Transcription Factors
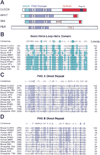
(A) The domain organization of CLOCK is similar to the original members of the PAS domain family—PERIOD, ARNT, and SIM. (B–D) The amino acid alignments of the basic–helix-loop-helix region and the PAS-A and PAS-B direct repeats of CLOCK and 10 other members of the bHLH–PAS domain family of transcription factors are shown (8 of these are mouse or human protein sequences; the remaining 2 are from Drosophila). PERIOD is included in the alignments of the PAS-A and PAS-B domains but lacks a basic–helix-loop-helix motif. Amino acid sequences were obtained through the Entrez sequence retrieval system (National Center for Biotechnology). Consensus amino acids represent residues identical among ≥50% of the proteins analyzed. Numbers in parentheses indicate position of the start of amino acid fragment with respect to the initiator methionine of the corresponding protein. NPAS2 shares the highest identity with CLOCK of all sequences in the database at the time of submission. There is very high sequence identity (>90%) in the basic portion of the basic–helix-loop-helix domain and the PAS B direct repeat. Accession numbers for the PAS proteins shown: Drosophila PER (PERIOD, S17286); mouse NPAS1 (Neuronal PAS1,U77967); mouseNPAS2 (NeuronalPAS2,U77969); Drosophila TRH (trachealess, U42699); human JAP3 (bHLH–PAS protein JAP3, U60415); mouse EPAS1 (endothelial PAS domain protein 1, U81983); Drosophila SIM (single minded, 134494); mouse AHR (Aryl hydrocarbon receptor, M94623); mouse SIM1 (mSim1, D79209); ARNT (Aryl Hydrocarbon Nuclear Translocator, M69238); human HIF_a_ (hypoxia-inducible factor 1 alpha, U22431).
Figure 5. Identification and Analysis of the Clock Mutation
(A) Locomotor activity records and DNA sequence data from wild-type and Clock/Clock isogenic C57BL/6J (B6) mice. Activity records are double plotted so that 48 hr are represented horizontally, and each 24 hr interval is presented both to the right of and beneath the preceding day. Wheel-running activity is indicated by black markings. Mice were kept on a light:dark cycle (LD 12:12) for the first 8 days shown, as indicated by the bar above each record. Beginning with lights-off on day 8, the animals were kept in continuous darkness for the remainder of the record. Shown below each activity record is the DNA sequence of the exon–intron junction 3′ to exon 19. The DNA used for sequencing was PCR amplified from the genomic DNA of the mice whose activity records are presented above each sequence trace. Arrows indicate the base that is altered in Clock/Clock mutant mice. This A→T transversion was the only nucleotide difference detected from a comparative sequence scan of all of the coding exons (and flanking intronic sequence) of this gene. (B) RT–PCR products. RNA samples for lanes 1 and 2 were reverse transcribed using random hexamer primers. RNA samples for lanes 3 and 4 were reverse transcribed using a gene-specific primer located in exon 23. PCR amplification was performed using gene specific primers from exons 15 and 21, which amplify a wild-type product of 761 bp. Lanes 1 and 3 are RT–PCR products amplified from hypothalamic RNA of wild-type isogenic B6 mice; lanes 2 and 4 are RT–PCR products amplified from hypothalamic RNA of Clock/Clock isogenic B6 mice. (C) Exon content of RT–PCR products. Sequencing of the RT–PCR products revealed that exon 19 is indeed missing in the RNA transcripts of Clock/Clock mice. The additional band seen in the RT–PCR products is the result of the variable splicing of exon 18.
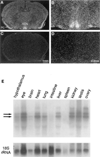
Figure 6. Analysis of Clock mRNA Expression
(A–D) In situ hybridization of Clock mRNA in the mouse brain. Coronal sections from brains of wild-type mice collected at ZT6, probed with yz50 antisense (A and B) and sense (C and D) ribo-probes. (E) Tissue distribution blot. Clock mRNA expression is not limited to the SCN. Northern blot analysis of total RNA extracted from diverse tissues (all of which were collected at ZT6 from wild type C57BL/6J mice) reveals that Clock expression is widespread. The bottom panel shows the same filter hybridized with a 32P-labeled oligonucleotide of 18S RNA for normalization.
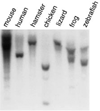
Figure 7. Evolutionary Conservation of the Clock Gene
Genomic DNA samples collected from mouse (C57BL/6J), human (D. P. K.), chicken, golden hamster (Mesocricetus auratus), lizard (Anolis carolinensis), frog (Xenopus laevis), and zebrafish (Danio rerio) were digested with PstI and analyzed by Southern blot using a mouse Clock PAS-B PCR probe (see Experimental Procedures). Similar results were obtained with a C-terminal probe (data not shown).
Similar articles
- Functional identification of the mouse circadian Clock gene by transgenic BAC rescue.
Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Antoch MP, et al. Cell. 1997 May 16;89(4):655-67. doi: 10.1016/s0092-8674(00)80246-9. Cell. 1997. PMID: 9160756 Free PMC article. - Molecular cloning and characterization of the human CLOCK gene: expression in the suprachiasmatic nuclei.
Steeves TD, King DP, Zhao Y, Sangoram AM, Du F, Bowcock AM, Moore RY, Takahashi JS. Steeves TD, et al. Genomics. 1999 Apr 15;57(2):189-200. doi: 10.1006/geno.1998.5675. Genomics. 1999. PMID: 10198158 - A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless.
Allada R, White NE, So WV, Hall JC, Rosbash M. Allada R, et al. Cell. 1998 May 29;93(5):791-804. doi: 10.1016/s0092-8674(00)81440-3. Cell. 1998. PMID: 9630223 - Genetics and neurobiology of circadian clocks in mammals.
Siepka SM, Yoo SH, Park J, Lee C, Takahashi JS. Siepka SM, et al. Cold Spring Harb Symp Quant Biol. 2007;72:251-259. doi: 10.1101/sqb.2007.72.052. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419282 Free PMC article. Review. - Clines in clock genes: fine-tuning circadian rhythms to the environment.
Kyriacou CP, Peixoto AA, Sandrelli F, Costa R, Tauber E. Kyriacou CP, et al. Trends Genet. 2008 Mar;24(3):124-32. doi: 10.1016/j.tig.2007.12.003. Epub 2008 Feb 19. Trends Genet. 2008. PMID: 18243399 Review.
Cited by
- Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms.
Lee IT, Chang AS, Manandhar M, Shan Y, Fan J, Izumo M, Ikeda Y, Motoike T, Dixon S, Seinfeld JE, Takahashi JS, Yanagisawa M. Lee IT, et al. Neuron. 2015 Mar 4;85(5):1086-102. doi: 10.1016/j.neuron.2015.02.006. Neuron. 2015. PMID: 25741729 Free PMC article. - How might circadian rhythms control mood? Let me count the ways..
McClung CA. McClung CA. Biol Psychiatry. 2013 Aug 15;74(4):242-9. doi: 10.1016/j.biopsych.2013.02.019. Epub 2013 Apr 1. Biol Psychiatry. 2013. PMID: 23558300 Free PMC article. Review. - Core circadian protein CLOCK is a positive regulator of NF-κB-mediated transcription.
Spengler ML, Kuropatwinski KK, Comas M, Gasparian AV, Fedtsova N, Gleiberman AS, Gitlin II, Artemicheva NM, Deluca KA, Gudkov AV, Antoch MP. Spengler ML, et al. Proc Natl Acad Sci U S A. 2012 Sep 11;109(37):E2457-65. doi: 10.1073/pnas.1206274109. Epub 2012 Aug 15. Proc Natl Acad Sci U S A. 2012. PMID: 22895791 Free PMC article. - Ammonia-lowering activities and carbamoyl phosphate synthetase 1 (Cps1) induction mechanism of a natural flavonoid.
Nohara K, Shin Y, Park N, Jeong K, He B, Koike N, Yoo SH, Chen Z. Nohara K, et al. Nutr Metab (Lond). 2015 Jun 9;12:23. doi: 10.1186/s12986-015-0020-7. eCollection 2015. Nutr Metab (Lond). 2015. PMID: 26075008 Free PMC article. - The Arg-293 of Cryptochrome1 is responsible for the allosteric regulation of CLOCK-CRY1 binding in circadian rhythm.
Gul S, Aydin C, Ozcan O, Gurkan B, Surme S, Baris I, Kavakli IH. Gul S, et al. J Biol Chem. 2020 Dec 11;295(50):17187-17199. doi: 10.1074/jbc.RA120.014333. Epub 2020 Oct 7. J Biol Chem. 2020. PMID: 33028638 Free PMC article.
References
- Akagi JM, Nomiyama H, Setoyama C, Akagi M. Messenger RNA expressed in mouse teratocarcinoma stem cells and down-regulated by a tumor-promoting phorbol ester codes for a novel transmembrane protein. Biochem. Biophys. Res. Commun. 1988;157:548–557. - PubMed
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J. Mol. Biol. 1990;215:403–410. - PubMed
- Aronson BD, Johnson KA, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. - PubMed
- Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 MH39592/MH/NIMH NIH HHS/United States
- T32 GM008152/GM/NIGMS NIH HHS/United States
- P30 HD28048/HD/NICHD NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM08152/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials