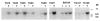Functional identification of the mouse circadian Clock gene by transgenic BAC rescue - PubMed (original) (raw)
Functional identification of the mouse circadian Clock gene by transgenic BAC rescue
M P Antoch et al. Cell. 1997.
Abstract
As a complementary approach to positional cloning, we used in vivo complementation with bacterial artificial chromosome (BAC) clones expressed in transgenic mice to identify the circadian Clock gene. A 140 kb BAC transgene completely rescued both the long period and the loss-of-rhythm phenotypes in Clock mutant mice. Analysis with overlapping BAC transgenes demonstrates that a large transcription unit spanning approximately 100,000 base pairs is the Clock gene and encodes a novel basic-helix-loop-helix-PAS domain protein. Overexpression of the Clock transgene can shorten period length beyond the wild-type range, which provides additional evidence that Clock is an integral component of the circadian pacemaking system. Taken together, these results provide a proof of principle that "cloning by rescue" is an efficient and definitive method in mice.
Figures
Figure 1
Physical Map of the Mouse Clock Locus Clock is localized to the mid-portion of mouse chromosome 5; SSLP markers D5Nwu2 and D5Mit307 define the nonrecombinant interval containing Clock. The relative positions of various sequence-tagged sites, SSLP markers, and restriction enzyme sites are indicated. D5Nwu2, D5Nwu1, D5Nwu14, D5Nwu7, and D5Nwu13 are SSLP markers described in King et al. (1997b). YAC55 appears to be a nonchimeric clone based on STS content mapping and long-range restriction mapping of overlapping YAC clones (data not shown). NotI, EagI, and NruI restriction profiles reveal two CpG islands. The three transcription units identified in this region, bendless, pFT27, and Clock, define the possible candidate genes; arrows indicate the direction of transcription and genomic extent, and introns and exons are indicated above the arrows. The BAC 54 rescuing clone is shaded. The two clones that fail to rescue, BAC 52 and the 100 kb NotI BAC 54 fragment, are shown as open bars. Telomeric (tel) and centromeric (cen) directions are indicated.
Figure 2
Transgene Copy Number Quantification BgIII-digested genomic DNA from transgenic and non-transgenic F1 progeny from each founder was probed with cDNA clone L4, which originates from the 3′untranslated region of the Clock gene. A 7kb fragment, which represents the endogenous Clock gene sequence) 1 copy/genome), was detected in all samples and was of much higher intensity in transgene-positive animals. A standard amount of BglII-digested BAC 54 DNA equivalent to 1, 3, and 5 copies per genome was mixed with 5 μg of control mouse DNA and loaded on the same gel. Band intensity was quantitated by densitometry, and transgene copy number was estimated (Table 1). (−) and (+) indicate mice negative or positive for transgene integration, respectively. The blot was normalized for DNA loading by hybridization with a mouse rhodopsin cDNA probe (a single copy copy gene [Al-Ubaidi et al., 1990]) (data not shown).
Figure 3
Genetic Cross Used to Produce Line TG36. Clock genotypes are depicted by shading, with wild-type open, heterozygotes shaded, and homozygotes closed. The presence of the 140kb BAC54 transgene is indicated by a double helix symbol. A Clock/+ transgenic founder male was bred with Clock heterozygous females so that mice of six genotypic classes (the three Clock genotypes, both with and without the transgene) were represented in the progeny.
Figure 4
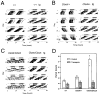
The BAC 54 Transgene Rescues Mutant Circadian Phenotype and Affects Wild-Type Circadian Activity Wheel running activity records and period estimates are shown for F1 transgenic and control mice from transgenic line TG36, which carries the BAC 54 transgene. All animals were maintained on an LD 12:12 cycle (as indicated by the light/dark bar) for 5–7 days and transferred into constant darkness at a time indicated by the arrow. (A) Activity records of the wild-type mice with (right) and without (left) the BAC 54 transgene (tg). (B) Activity records of _Clock/_+ mice with (right) and without (left) the BAC 54 transgene. (C) Activity records of Clock/Clock mice with (right) and without (left) the BAC 54 transgene. (D) Histogram of period estimates for 83 F1 progeny from line TG36. Individual data points are indicated by open circles. Not all individual data points can be seen due to their overlap. Numbers on the bottom of the bars indicate the number of animals in each group that were wheel tested for their circadian behavior. Within this line, significant effects of Clock genotype (2 DF, F = 239, p < 0.00001), transgene presence (1 DF, F = 645, p < 0.00001), as well as genotype by transgene interactions (2 DF, F = 205, p < 0.00001) were detected (GLM ANOVA). Wild-type transgenic mice had significantly shorter periods than all non-transgenic groups, Clock/+ and Clock/Clock transgenic mice had significantly shorter periods than Clock/+ and Clock/Clock non-transgenic mice, and Clock/Clock non-transgenic mice had significantly longer periods than all other groups as determined by Tukey’s post hoc tests (p < 0.05).
Figure 5
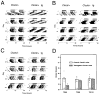
The 100 kb BAC 54 and BAC 52 Transgenes Do Not Affect Circadian Phenotype Wheel running activity records and period estimates of Clock/+ F1 progeny from different BAC transgenic lines are shown. Both transgenic and control animals were behaviorally tested using the experimental protocol described in Figure 4 and the text. (A) Activity records of line TG55 Clock/+ progeny, in which the full-length BAC 54 was the transgenic construct. (tg) indicates transgenic animals’ records. (B) Activity records of line TG80 Clock/+ progeny, in which the 100 kb BAC 54 fragment was the transgenic construct. (C) Activity records of line TG121 Clock/+ progeny, in which the full-length BAC 52 was the transgenic construct. (D) Histogram of period estimates for 46 F1 animals from lines TG55, TG80, and TG121. Individual data points are indicated by open circles. Not all individual points can be seen due to overlapping values. Number of animals from each line tested is indicated on the bottom of the bar. Among the Clock/+ mice from these lines, significant effects of transgenic construct (2 DF, F = 9.60, p < 0.0005), transgene presence (1DF, F = 16.55, p < 0.0005), as well as construct by transgene interactions (2 DF, F = 10.8, p < 0.0001) were detected (GLM ANOVA). TG55 transgenic progeny had significantly shorter periods than all other groups as determined by Tukey’s post hoc tests (p < 0.05). There were no significant differences between transgenic and non-transgenic Clock/+ progeny in lines TG80 and TG121.
Figure 6
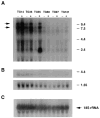
Clock mRNA Expression in Rescuing and Non-rescuing Lines Total RNA was isolated from the hypothalamic tissue of different BAC transgenic and control mice for Northern blot hybridization. Total RNA (25 μg) was hybridized with 32P-labeled 51-base oligonucleotide complementary to 18S ribosomal RNA (C). RNA loading differs across the lines, as can be seen with the 18S ribosomal probe; however, the critical pairwise comparisons between non-transgenic (−) and transgenic (+) mice within each line can still be made. The BAC 54 transgenic lines TG14, TG36, and TG55 showed increased levels of Clock mRNA expression in transgene-positive mice, whereas the 100 kb BAC 54 line (TG97) and the BAC 52 line (TG121) showed similar levels of Clock mRNA between non-transgenic and transgenic mice. In this experiment, increased expression of Clock mRNA was not apparent in the BAC 54 line TG60 due to the low copy number of the transgene. Transgenic mice from the TG55 line also expressed a novel 2.4 kb transcript.
Figure 7
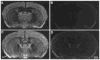
In Situ Hybridization Analysis of Clock mRNA in Line TG36 Coronal brain sections (20 μm) from TG36 Clock/Clock littermates, either lacking the transgene (A and B) or positive for the transgene (C and D), are shown. (A) and (C) are autoradiograms using antisense yz50probe, while (B) and (D) show the sense probe controls. Expression of Clock mRNA is widely distributed in the brains of both non-transgenic and transgenic animals. Highest levels of Clock mRNA were found in the suprachiasmatic nucleus (SCN) and pyriform cortex (PC). The transgene-positive animal (C) shows a selectively stronger signal in SCN as compared to Clock/Clock control animal (A). These representative sections are from the midportion of the SCN.
Similar articles
- Positional cloning of the mouse circadian clock gene.
King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. King DP, et al. Cell. 1997 May 16;89(4):641-53. doi: 10.1016/s0092-8674(00)80245-7. Cell. 1997. PMID: 9160755 Free PMC article. - The mouse Clock locus: sequence and comparative analysis of 204 kb from mouse chromosome 5.
Wilsbacher LD, Sangoram AM, Antoch MP, Takahashi JS. Wilsbacher LD, et al. Genome Res. 2000 Dec;10(12):1928-40. doi: 10.1101/gr.10.12.1928. Genome Res. 2000. PMID: 11116088 Free PMC article. - Genetics and neurobiology of circadian clocks in mammals.
Siepka SM, Yoo SH, Park J, Lee C, Takahashi JS. Siepka SM, et al. Cold Spring Harb Symp Quant Biol. 2007;72:251-259. doi: 10.1101/sqb.2007.72.052. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419282 Free PMC article. Review. - Molecular cloning and characterization of the human CLOCK gene: expression in the suprachiasmatic nuclei.
Steeves TD, King DP, Zhao Y, Sangoram AM, Du F, Bowcock AM, Moore RY, Takahashi JS. Steeves TD, et al. Genomics. 1999 Apr 15;57(2):189-200. doi: 10.1006/geno.1998.5675. Genomics. 1999. PMID: 10198158 - Oscillating perceptions: the ups and downs of the CLOCK protein in the mouse circadian system.
Debruyne JP. Debruyne JP. J Genet. 2008 Dec;87(5):437-46. doi: 10.1007/s12041-008-0066-7. J Genet. 2008. PMID: 19147932 Free PMC article. Review.
Cited by
- Methylation of histone H3 on lysine 4 by the lysine methyltransferase SET1 protein is needed for normal clock gene expression.
Raduwan H, Isola AL, Belden WJ. Raduwan H, et al. J Biol Chem. 2013 Mar 22;288(12):8380-8390. doi: 10.1074/jbc.M112.359935. Epub 2013 Jan 14. J Biol Chem. 2013. PMID: 23319591 Free PMC article. - Manipulating the circadian and sleep cycles to protect against metabolic disease.
Nohara K, Yoo SH, Chen ZJ. Nohara K, et al. Front Endocrinol (Lausanne). 2015 Mar 23;6:35. doi: 10.3389/fendo.2015.00035. eCollection 2015. Front Endocrinol (Lausanne). 2015. PMID: 25852644 Free PMC article. Review. - Small molecule modifiers of circadian clocks.
Chen Z, Yoo SH, Takahashi JS. Chen Z, et al. Cell Mol Life Sci. 2013 Aug;70(16):2985-98. doi: 10.1007/s00018-012-1207-y. Epub 2012 Nov 16. Cell Mol Life Sci. 2013. PMID: 23161063 Free PMC article. Review. - Mutations in Mlph, encoding a member of the Rab effector family, cause the melanosome transport defects observed in leaden mice.
Matesic LE, Yip R, Reuss AE, Swing DA, O'Sullivan TN, Fletcher CF, Copeland NG, Jenkins NA. Matesic LE, et al. Proc Natl Acad Sci U S A. 2001 Aug 28;98(18):10238-43. doi: 10.1073/pnas.181336698. Epub 2001 Aug 14. Proc Natl Acad Sci U S A. 2001. PMID: 11504925 Free PMC article. - A mutation in Rab27a causes the vesicle transport defects observed in ashen mice.
Wilson SM, Yip R, Swing DA, O'Sullivan TN, Zhang Y, Novak EK, Swank RT, Russell LB, Copeland NG, Jenkins NA. Wilson SM, et al. Proc Natl Acad Sci U S A. 2000 Jul 5;97(14):7933-8. doi: 10.1073/pnas.140212797. Proc Natl Acad Sci U S A. 2000. PMID: 10859366 Free PMC article.
References
- Akagi JM, Nomiyama H, Setoyama C, Akagi M. Messenger RNA expressed in mouse teratocarcinoma stem cells and down-regulated by a tumor-promoting phorbol ester codes for a novel transmembrane protein. Biochem Biophys Res Commun. 1988;157:548–557. - PubMed
- Al-Ubaidi MR, Pittler SJ, Champagne MS, Triantafyllos JT, McGinnis JF, Baehr W. Mouse opsin: gene structure and molecular basis of multiple transcripts. J Biol Chem. 1990;265:20563–20569. - PubMed
- Aronson BD, Johnson KA, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. - PubMed
- Bargiello TA, Jackson FR, Young MW. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature. 1984;312:752–754. - PubMed
- Baylies MK, Bargiello TA, Jackson FR, Young MW. Changes in abundance or structure of the per gene product can alter periodicity of the Drosophila clock. Nature. 1987;326:390–392. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 MH39592/MH/NIMH NIH HHS/United States
- T32 GM08152/GM/NIGMS NIH HHS/United States
- T32 GM008152/GM/NIGMS NIH HHS/United States
- P30 HD28048/HD/NICHD NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials