Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C - PubMed (original) (raw)
Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C
Q Lin et al. Science. 1997.
Abstract
Members of the myocyte enhancer factor-2 (MEF2) family of MADS (MCM1, agamous, deficiens, serum response factor)-box transcription factors bind an A-T-rich DNA sequence associated with muscle-specific genes. The murine MEF2C gene is expressed in heart precursor cells before formation of the linear heart tube. In mice homozygous for a null mutation of MEF2C, the heart tube did not undergo looping morphogenesis, the future right ventricle did not form, and a subset of cardiac muscle genes was not expressed. The absence of the right ventricular region of the mutant heart correlated with down-regulation of the dHAND gene, which encodes a basic helix-loop-helix transcription factor required for cardiac morphogenesis. Thus, MEF2C is an essential regulator of cardiac myogenesis and right ventricular development.
Figures
Fig. 1
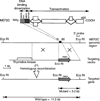
Targeting strategy for the MEF2C locus. Diagram of MEF2C, which contains 465 amino acids, is shown at the top, above a diagram of the mouse MEF2C locus around coding exon 2, which encodes residues 18 to 86. The first intron in the coding region is >10 kb. In the targeting vector (10), the neomycin-resistance gene (neo) was transcribed in the same direction as MEF2C. Homologous recombination resulted in deletion of coding exon 2. The structure of the targeted allele is shown at the bottom. Insertion of neo introduced an Eco RI site that could be used to distinguish the wild-type and mutant alleles. Position of the 3′ probe used for Southern blot analysis of yolk sac DNA is indicated. Hybridization of Eco RI– digested DNA with the 3′ probe yielded fragments of 11.5 and 3.2 kb from the wild-type and mutant loci, respectively.
Fig. 2
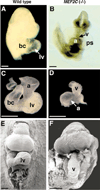
Cardiac defects in MEF2C null embryos. (A) Wild-type and (B) mutant embryos at E9.0. The lower portion of the wild-type embryo was removed for a clear view of the heart. Note severe pericardial effusion in the mutant. Hearts of the wild-type (C) and mutant (D) embryos are shown. Scanning electron micrographs of wild-type (E) and mutant (F) embryos at E9.0. Abbreviations: a, Atrium; bc, bulbus cordis; lv, future left ventricle; ps, pericardial sac; and v, ventricular chamber. Bars = 2 µm.
Fig. 3
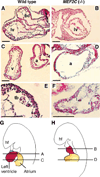
Hematoxylin and eosin staining of thin sections of hearts of wild-type and mutant embryos. Wild-type (A, C, and E) and mutant (B, D, and F) embryos at E9.0 were fixed, sectioned, and stained with hematoxylin and eosin (27). (G and H) Diagrams of planes of section. Abbreviations: a, Atrium; bc, bulbus cordis; hf, headfold; lv, left ventricle; tb, trabeculae; and v, ventricular chamber. Bars = 1 µm.
Fig. 4
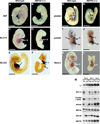
Analysis of cardiac gene expression by whole mount in situ hybridization. Wild-type (A, C, E, G, I, and K) and mutant (B, D, F, H, J, and L) embryos were stained by in situ hybridization for the indicated transcripts (3, 28). For a clearer view of the heart, heads and tails were dissected away from the embryos in (I), (J), and (L). Note that dHAND expression is restricted to the right ventricular region of the heart in the wild-type embryo and is not expressed in the heart of the mutant. eHAND is expressed in the conotruncus and left ventricular region in the wild-type heart; in the mutant, there is no gap in expression that would represent the right ventricle. Tails were dissected away from embryos in (C), (D), and (K). (F), (I), (J), and (L) are frontal views; other embryos are viewed from the side. Abbreviations: a, Atrium; ct, conotruncus; h, heart; lm, lateral mesoderm; lv, left ventricle; rv, right ventricle; and v, ventricular chamber. Bars = 5 µm. (M) Quantitative RT-PCR analysis of gene expression in wild-type (wt) and MEF2C mutant (mu) embryos. PCR amplifications were performed with cDNA synthesized from RNA isolated from E9.5 embryonic heart or whole embryos at E8.5 and E9.0 and gene-specific primers for the indicated transcripts. L7 was used as a loading control. Transcripts for dHAND were down-regulated in the heart by E9.0, as determined by in situ hybridization. The expression of dHAND transcripts in the E8.5 and E9.0 RNA samples used for RT-PCR reflects its expression in the lateral mesoderm and not in the heart. RNA concentrations were quantitated by PhosphorImager analysis.
Similar articles
- mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo.
Dodou E, Xu SM, Black BL. Dodou E, et al. Mech Dev. 2003 Sep;120(9):1021-32. doi: 10.1016/s0925-4773(03)00178-3. Mech Dev. 2003. PMID: 14550531 - Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis.
Edmondson DG, Lyons GE, Martin JF, Olson EN. Edmondson DG, et al. Development. 1994 May;120(5):1251-63. doi: 10.1242/dev.120.5.1251. Development. 1994. PMID: 8026334 - Disruption of MEF2 activity in cardiomyoblasts inhibits cardiomyogenesis.
Karamboulas C, Dakubo GD, Liu J, De Repentigny Y, Yutzey K, Wallace VA, Kothary R, Skerjanc IS. Karamboulas C, et al. J Cell Sci. 2006 Oct 15;119(Pt 20):4315-21. doi: 10.1242/jcs.03186. Epub 2006 Sep 26. J Cell Sci. 2006. PMID: 17003108 - Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins.
Black BL, Olson EN. Black BL, et al. Annu Rev Cell Dev Biol. 1998;14:167-96. doi: 10.1146/annurev.cellbio.14.1.167. Annu Rev Cell Dev Biol. 1998. PMID: 9891782 Review. - Seeking a regulatory roadmap for heart morphogenesis.
Harvey RP. Harvey RP. Semin Cell Dev Biol. 1999 Feb;10(1):99-107. doi: 10.1006/scdb.1998.0277. Semin Cell Dev Biol. 1999. PMID: 10355034 Review.
Cited by
- 12-O-Tetradecanoylphorbol-13-acetate increases cardiomyogenesis through PKC/ERK signaling.
Radaszkiewicz KA, Beckerová D, Woloszczuková L, Radaszkiewicz TW, Lesáková P, Blanářová OV, Kubala L, Humpolíček P, Pachernik J. Radaszkiewicz KA, et al. Sci Rep. 2020 Sep 28;10(1):15922. doi: 10.1038/s41598-020-73074-4. Sci Rep. 2020. PMID: 32985604 Free PMC article. - Variable paralog expression underlies phenotype variation.
Bailon-Zambrano R, Sucharov J, Mumme-Monheit A, Murry M, Stenzel A, Pulvino AT, Mitchell JM, Colborn KL, Nichols JT. Bailon-Zambrano R, et al. Elife. 2022 Sep 22;11:e79247. doi: 10.7554/eLife.79247. Elife. 2022. PMID: 36134886 Free PMC article. - Structure and regulation of human troponin genes.
Cullen ME, Dellow KA, Barton PJ. Cullen ME, et al. Mol Cell Biochem. 2004 Aug;263(1):81-90. doi: 10.1023/B:MCBI.0000041850.37415.b8. Mol Cell Biochem. 2004. PMID: 27520667 - The NADPH oxidase NOX4 drives cardiac differentiation: Role in regulating cardiac transcription factors and MAP kinase activation.
Li J, Stouffs M, Serrander L, Banfi B, Bettiol E, Charnay Y, Steger K, Krause KH, Jaconi ME. Li J, et al. Mol Biol Cell. 2006 Sep;17(9):3978-88. doi: 10.1091/mbc.e05-06-0532. Epub 2006 Jun 14. Mol Biol Cell. 2006. PMID: 16775014 Free PMC article. - Atrial AMP-activated protein kinase is critical for prevention of dysregulation of electrical excitability and atrial fibrillation.
Su KN, Ma Y, Cacheux M, Ilkan Z, Raad N, Muller GK, Wu X, Guerrera N, Thorn SL, Sinusas AJ, Foretz M, Viollet B, Akar JG, Akar FG, Young LH. Su KN, et al. JCI Insight. 2022 Apr 22;7(8):e141213. doi: 10.1172/jci.insight.141213. JCI Insight. 2022. PMID: 35451373 Free PMC article.
References
- Edmondson DG, Lyons GE, Martin JF, Olson EN. Development. 1994;120:1251. - PubMed
- Olson EN, Perry M, Schulz RA. Dev. Biol. 1995;172:280. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases