STRL33, A novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1 - PubMed (original) (raw)
STRL33, A novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1
F Liao et al. J Exp Med. 1997.
Abstract
The chemokine receptors CXCR4, CCR2B, CCR3, and CCR5 have recently been shown to serve along with CD4 as coreceptors for HIV-1. The tropisms of HIV-1 strains for subgroups of CD4(+) cells can be explained, at least partly, by the selective use of G protein-coupled receptors (GPCRs). We have identified a novel human gene, STRL33, located on chromosome 3 that encodes a GPCR with sequence similarity to chemokine receptors and to chemokine receptor-like orphan receptors. STRL33 is expressed in lymphoid tissues and activated T cells, and is induced in activated peripheral blood lymphocytes. When transfected into nonhuman NIH 3T3 cells expressing human CD4, the STRL33 cDNA rendered these cells competent to fuse with cells expressing HIV-1 envelope glycoproteins (Envs). Of greatest interest, STRL33, in contrast with CXCR4 or CCR5, was able to function as a cofactor for fusion mediated by Envs from both T cell line-tropic and macrophage-tropic HIV-1 strains. STRL33-transfected Jurkat cell lines also supported enhanced productive infection with HIV-1 compared with control Jurkat cells. Despite the sequence similarities between STRL33 and chemokine receptors, STRL33-transfected cell lines did not respond to any in a panel of chemokines. Based on the pattern of tissue expression of the STRL33 mRNA, and given the ability of STRL33 to function with Envs of differing tropisms, STRL33 may play a role in the establishment and/or progression of HIV-1 infection.
Figures
Figure 1
Alignment of the STRL33 predicted amino acid sequence with the selected GPCRs STRL22, GPR-9-6, EBI1, IL-8RB, CXCR4, CCR3, CCR5, and IL-8RA. Numbers at the right indicate the positions of the residues at the end of each line of sequence. Solid backgrounds highlight matches between STRL33 and the other receptors. Dots indicate gaps introduced for optimal alignments. Putative TMDs I–VII are indicated by bars. The alignments were generated using the PileUp program of the Wisconsin Sequence Analysis Package of Genetics Computer Group (Madison, WI).
Figure 2
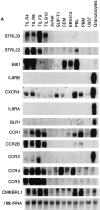
Expression of STRL33 and other GPCR genes. (A) The expression of STRL33, genes for known chemokine receptors, and genes for selected orphan GPCRs in leukocytes. 15 μg of total RNA were electrophoresed on 1.2% agarose–formaldehyde gels, transferred to nitrocellulose membranes, and hybridized with the probes indicated on the left. A total of six membranes were used for hybridizations, and adequate removal of signal was documented before repeat probings. Film exposure times ranged from overnight for the IL-8RA and IL-8RB blots to 13 d for the CXCR4 blot. Probings were done using an oligonucleotide complementary to 18S rRNA in order to demonstrate amounts of RNA loaded per lane and a representative blot is shown. (B) The expression of STRL33 in activated PBL. 25 μg of total RNA from TIL, and freshly isolated and activated PBL were analyzed as in A and hybridized with a 32P-labeled STRL33 ORF probe and a probe for 18S rRNA.
Figure 2
Expression of STRL33 and other GPCR genes. (A) The expression of STRL33, genes for known chemokine receptors, and genes for selected orphan GPCRs in leukocytes. 15 μg of total RNA were electrophoresed on 1.2% agarose–formaldehyde gels, transferred to nitrocellulose membranes, and hybridized with the probes indicated on the left. A total of six membranes were used for hybridizations, and adequate removal of signal was documented before repeat probings. Film exposure times ranged from overnight for the IL-8RA and IL-8RB blots to 13 d for the CXCR4 blot. Probings were done using an oligonucleotide complementary to 18S rRNA in order to demonstrate amounts of RNA loaded per lane and a representative blot is shown. (B) The expression of STRL33 in activated PBL. 25 μg of total RNA from TIL, and freshly isolated and activated PBL were analyzed as in A and hybridized with a 32P-labeled STRL33 ORF probe and a probe for 18S rRNA.
Figure 3
The expression of STRL33 in human tissues. Blots were prepared by the supplier (Clontech, Palo Alto, CA) from 1.2% agarose–formaldehyde gels containing ∼2 μg poly(A)+ RNA per lane. Hybridizations were done using a 32P-labeled STRL33 ORF probe and blots were washed according to the instructions of the manufacturer. The blot prepared from lymphoid tissue (left) was exposed for 2 d, and the blot from other selected tissues (right) was exposed for 8 d.
Figure 4
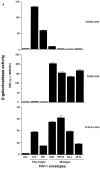
Activity of STRL33 as a fusion cofactor. (A) NIH 3T3 cells were transfected with DNAs encoding CXCR4 or CCR5 or STRL33, and infected with vaccinia recombinants encoding CD4 and T7 RNA polymerase. HeLa cells were infected with a vaccinia recombinant containing lacZ under control of a T7 promoter and infected separately with vaccinia recombinants encoding the indicated Envs. Unc is a mutant Env that cannot be cleaved to gp120 and gp41 and cannot mediate fusion. Cell fusion was quantified by measuring β-Gal activity. NIH 3T3 cells transfected with the STRL33 cDNA but not infected with virus vCB-3 encoding CD4 did not fuse with cells expressing any of the Envs (data not shown). Results of one experiment are shown. STRL33 also mediated fusion with cells expressing both TCL-tropic and M-tropic Envs in four additional experiments. (B) Cell fusion experiment done as in A, except that here the DNA encoding the Env, 89.6, was introduced into HeLa cells by transfection of plasmid DNA, resulting in lower levels of fusion overall as compared with A. The value obtained for Unc has been subtracted from each of the values obtained using the 89.6 Env. Results are shown from one experiment. Similar activity for STRL33 was found in another experiment using the 89.6 Env.
Figure 4
Activity of STRL33 as a fusion cofactor. (A) NIH 3T3 cells were transfected with DNAs encoding CXCR4 or CCR5 or STRL33, and infected with vaccinia recombinants encoding CD4 and T7 RNA polymerase. HeLa cells were infected with a vaccinia recombinant containing lacZ under control of a T7 promoter and infected separately with vaccinia recombinants encoding the indicated Envs. Unc is a mutant Env that cannot be cleaved to gp120 and gp41 and cannot mediate fusion. Cell fusion was quantified by measuring β-Gal activity. NIH 3T3 cells transfected with the STRL33 cDNA but not infected with virus vCB-3 encoding CD4 did not fuse with cells expressing any of the Envs (data not shown). Results of one experiment are shown. STRL33 also mediated fusion with cells expressing both TCL-tropic and M-tropic Envs in four additional experiments. (B) Cell fusion experiment done as in A, except that here the DNA encoding the Env, 89.6, was introduced into HeLa cells by transfection of plasmid DNA, resulting in lower levels of fusion overall as compared with A. The value obtained for Unc has been subtracted from each of the values obtained using the 89.6 Env. Results are shown from one experiment. Similar activity for STRL33 was found in another experiment using the 89.6 Env.
Figure 5
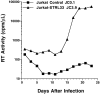
HIV-1 infection of _STRL33_-transfected Jurkat cells. 2 × 106 cells were infected with HIV-1ELI1 corresponding to 105 cpm of RT activity. Samples were taken every 2 d for determining RT activity from cultures of the control-transfected Jurkat cell line JC0.1 (▪) and from cultures of the _STRL33._1-transfected Jurkat cell line JC3.9 (▴).
Similar articles
- CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1.
Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. Alkhatib G, et al. Science. 1996 Jun 28;272(5270):1955-8. doi: 10.1126/science.272.5270.1955. Science. 1996. PMID: 8658171 - Coreceptor usage of BOB/GPR15 and Bonzo/STRL33 by primary isolates of human immunodeficiency virus type 1.
P Hlmann S, Krumbiegel M, Kirchhoff F. P Hlmann S, et al. J Gen Virol. 1999 May;80 ( Pt 5):1241-1251. doi: 10.1099/0022-1317-80-5-1241. J Gen Virol. 1999. PMID: 10355771 - Human chemokine receptors CCR5, CCR3 and CCR2B share common polarity motif in the first extracellular loop with other human G-protein coupled receptors implications for HIV-1 coreceptor function.
Efremov R, Truong MJ, Darcissac EC, Zeng J, Grau O, Vergoten G, Debard C, Capron A, Bahr GM. Efremov R, et al. Eur J Biochem. 1999 Aug;263(3):746-56. doi: 10.1046/j.1432-1327.1999.00553.x. Eur J Biochem. 1999. PMID: 10469138 - Chemokine receptors and HIV-1: the fusion of two major research fields.
Horuk R. Horuk R. Immunol Today. 1999 Feb;20(2):89-94. doi: 10.1016/s0167-5699(98)01396-6. Immunol Today. 1999. PMID: 10098328 Review. - Chemokine receptors and HIV.
Broder CC, Collman RG. Broder CC, et al. J Leukoc Biol. 1997 Jul;62(1):20-9. doi: 10.1002/jlb.62.1.20. J Leukoc Biol. 1997. PMID: 9225988 Review.
Cited by
- Virus-Host Protein Interaction Network of the Hepatitis E Virus ORF2-4 by Mammalian Two-Hybrid Assays.
Corneillie L, Lemmens I, Weening K, De Meyer A, Van Houtte F, Tavernier J, Meuleman P. Corneillie L, et al. Viruses. 2023 Dec 12;15(12):2412. doi: 10.3390/v15122412. Viruses. 2023. PMID: 38140653 Free PMC article. - Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1 gp120 involved in CCR5 utilization.
Hung CS, Vander Heyden N, Ratner L. Hung CS, et al. J Virol. 1999 Oct;73(10):8216-26. doi: 10.1128/JVI.73.10.8216-8226.1999. J Virol. 1999. PMID: 10482572 Free PMC article. - Determination of coreceptor usage of human immunodeficiency virus type 1 from patient plasma samples by using a recombinant phenotypic assay.
Trouplin V, Salvatori F, Cappello F, Obry V, Brelot A, Heveker N, Alizon M, Scarlatti G, Clavel F, Mammano F. Trouplin V, et al. J Virol. 2001 Jan;75(1):251-9. doi: 10.1128/JVI.75.1.251-259.2001. J Virol. 2001. PMID: 11119595 Free PMC article. - Potential contributions of viral envelope and host genetic factors in a human immunodeficiency virus type 1-infected long-term survivor.
Grovit-Ferbas K, Ferbas J, Gudeman V, Sadeghi S, Goetz MB, Giorgi JV, Chen IS, O'Brien WA. Grovit-Ferbas K, et al. J Virol. 1998 Nov;72(11):8650-8. doi: 10.1128/JVI.72.11.8650-8658.1998. J Virol. 1998. PMID: 9765405 Free PMC article. - Bicyclams, selective antagonists of the human chemokine receptor CXCR4, potently inhibit feline immunodeficiency virus replication.
Egberink HF, De Clercq E, Van Vliet AL, Balzarini J, Bridger GJ, Henson G, Horzinek MC, Schols D. Egberink HF, et al. J Virol. 1999 Aug;73(8):6346-52. doi: 10.1128/JVI.73.8.6346-6352.1999. J Virol. 1999. PMID: 10400726 Free PMC article.
References
- Murphy P. The molecular biology of leukocyte chemoattractant receptors. Annu Rev Immunol. 1994;12:593–633. - PubMed
- Schall TJ, Bacon K. Chemokines, leukocyte trafficking and inflammation. Curr Opin Immunol. 1994;6:865–873. - PubMed
- Raport CJ, Schweickart VL, Chantry D, Eddy RLJ, Shows T, Godiska R, Gray PW. New members of the chemokine receptor gene family. J Leuk Biol. 1996;59:18–23. - PubMed
- Miedema F, Meyaard C, Koot M, Klein MR, Roos MT, Groenink M, Fouchier RA, Van't Wout A. Changing virus–host interactions in the course of HIV-1 infection. Immunol Rev. 1994;140:35–72. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials