Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis - PubMed (original) (raw)
Comparative Study
Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis
D G Oppenheimer et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Little is known about how cell shape is controlled. We are using the morphogenesis of trichomes (plant hairs) on the plant Arabidopsis thaliana as a model to study how cell shape is controlled. Wild-type Arabidopsis trichomes are large, single epidermal cells with a stalk and three or four branches, whereas in zwichel (zwi) mutants the trichomes have a shortened stalk and only two branches. To further understand the role of the ZWI gene in trichome morphogenesis we have cloned the wild-type ZWICHEL (ZWI) gene by T-DNA tagging, and report here that it encodes a member of the kinesin superfamily of microtubule motor proteins. Kinesin proteins transport diverse cellular materials in a directional manner along microtubules. Kinesin-like proteins are characterized by a highly conserved "head" region that comprises the motor domain, and a nonconserved "tail" region that is thought to participate in recognition and binding of the appropriate cargo.
Figures
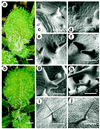
Figure 1
Development of wild-type and mutant trichomes. (A and B) Light micrographs of wild-type (A) and _zwi_-3 mutant (ecotype Columbia) (B) plants showing the distribution and shape of the trichomes. (C_–_J) Scanning electron micrographs of wild-type (C, E, G, and I) and _zwi_-3 mutant (D, F, H, and J) trichomes at different stages of development. Open arrow indicates distal tip of the leaf blade. Closed arrow in C indicates the second focus of cell expansion, which produces the first branch. [Scale bars = 1 mm (A and B), 43 μm (C_–_G), 86 μm (H), and 250 μm (I and J).]
Figure 2
Map of the ZWI locus. Open boxes represent the regions of the locus used in mutant rescue (complementation) experiments. The ability of the region to rescue the mutant phenotype is indicated on the left. At least four independent transformants were scored for each fragment. The shaded triangles represent the location of the T-DNA insertions. The open triangle represents the approximate location of the fast neutron-induced insertion. The diagram shown below the genomic region indicates the positions of the introns and exons. Exons are represented by open boxes, introns by solid boxes, and nontranslated regions by a solid line.
Figure 3
Complementation of mutant phenotype by transformation. Light micrograph of two independent regenerating transformants showing wild-type trichome morphology. These explants were transformed by Agrobacterium strain AGL1 carrying the subclone indicated in Fig. 2. (Scale bars = 1 mm.)
Figure 4
ZWI protein comparisons. (A) Comparison of the putative motor domain of the ZWI protein with the motor domains of other kinesin-like proteins. Dots represent amino acids identical to the ZWI sequence; dashes represent gaps that have been introduced to improve the alignment. The putative ATP-binding and microtubule-binding regions are indicated. KATA, Arabidopsis kinesin-like protein (22); KAR3, Saccharomyces kinesin-like protein (23); CLAR, Drosophila ncd+ (24); KIN, Drosophila kinesin heavy chain (25). (B) Comparison of the region of similarity between the Class IV Acanthamoeba myosin and ZWI. Dots in the protein sequence represent gaps that have been introduced to give optimal alignment. Vertical lines indicate amino acids that are identical, and colons represent similar amino acids. The numbers indicate amino acid positions. (C) Deduced amino acid sequence of the ZWI protein. Solid triangles show the positions of the introns. The open triangle shows the position of one of the T-DNA insertions. (The other T-DNA insertion occurs outside of the coding sequence.) The region with homology to the Acanthamoeba myosin is shown in boldface type, the kinesin-like motor domain is boxed, and the calmodulin-binding region is underlined.
Similar articles
- BEACH domain-containing protein SPIRRIG facilitates microtubule cytoskeleton-associated trichome morphogenesis in Arabidopsis.
Niu L, Xie W, Li Q, Wang Y, Zhang X, Shi M, Zeng J, Li M, Wang Y, Shao J, Yu F, An L. Niu L, et al. Planta. 2024 Oct 14;260(5):115. doi: 10.1007/s00425-024-04545-5. Planta. 2024. PMID: 39400709 - Extragenic suppressors of the arabidopsis zwi-3 mutation identify new genes that function in trichome branch formation and pollen tube growth.
Krishnakumar S, Oppenheimer DG. Krishnakumar S, et al. Development. 1999 Jun;126(14):3079-88. doi: 10.1242/dev.126.14.3079. Development. 1999. PMID: 10375500 - Developmental and cell-specific expression of ZWICHEL is regulated by the intron and exon sequences of its gene.
Reddy VS, Reddy AS. Reddy VS, et al. Plant Mol Biol. 2004 Jan;54(2):273-93. doi: 10.1023/B:PLAN.0000028793.88757.8b. Plant Mol Biol. 2004. PMID: 15159628 - Genetics of plant cell shape.
Oppenheimer DG. Oppenheimer DG. Curr Opin Plant Biol. 1998 Dec;1(6):520-4. doi: 10.1016/s1369-5266(98)80045-9. Curr Opin Plant Biol. 1998. PMID: 10066632 Review. - How plants split hairs.
Hülskamp M. Hülskamp M. Curr Biol. 2000 Apr 20;10(8):R308-10. doi: 10.1016/s0960-9822(00)00437-1. Curr Biol. 2000. PMID: 10801408 Review.
Cited by
- Plant Kinesin Repertoires Expand with New Domain Architecture and Contract with the Loss of Flagella.
Lucas J, Geisler M. Lucas J, et al. J Mol Evol. 2024 Aug;92(4):381-401. doi: 10.1007/s00239-024-10178-9. Epub 2024 Jun 26. J Mol Evol. 2024. PMID: 38926179 - The novel functions of kinesin motor proteins in plants.
Li J, Xu Y, Chong K. Li J, et al. Protoplasma. 2012 Jun;249 Suppl 2(Suppl 2):S95-100. doi: 10.1007/s00709-011-0357-3. Epub 2011 Dec 14. Protoplasma. 2012. PMID: 22167300 Free PMC article. Review. - Cytoskeleton and plant organogenesis.
Kost B, Bao YQ, Chua NH. Kost B, et al. Philos Trans R Soc Lond B Biol Sci. 2002 Jun 29;357(1422):777-89. doi: 10.1098/rstb.2002.1090. Philos Trans R Soc Lond B Biol Sci. 2002. PMID: 12079673 Free PMC article. Review. - Unraveling the lncRNA-miRNA-mRNA Regulatory Network Involved in Poplar Coma Development through High-Throughput Sequencing.
Song Z, Zhang C, Song G, Wei H, Xu W, Pan H, Ding C, Xu M, Zhen Y. Song Z, et al. Int J Mol Sci. 2024 Jul 5;25(13):7403. doi: 10.3390/ijms25137403. Int J Mol Sci. 2024. PMID: 39000510 Free PMC article. - IRREGULAR TRICHOME BRANCH1 in Arabidopsis encodes a plant homolog of the actin-related protein2/3 complex activator Scar/WAVE that regulates actin and microtubule organization.
Zhang X, Dyachok J, Krishnakumar S, Smith LG, Oppenheimer DG. Zhang X, et al. Plant Cell. 2005 Aug;17(8):2314-26. doi: 10.1105/tpc.104.028670. Epub 2005 Jul 8. Plant Cell. 2005. PMID: 16006582 Free PMC article.
References
- Cleland R E. In: Physiology of Cell Expansion During Plant Growth. Cosgrove D J, Knievel D P, editors. Rockville, MD: Am. Soc. Plant Physiol.; 1987. pp. 18–27.
- Cosgrove D J. Int J Plant Sci. 1993;154:10–21. - PubMed
- Ray P M. In: Physiology of Cell Expansion During Plant Growth. Cosgrove D J, Knievel D P, editors. Rockville, MD: Am. Soc. Plant Physiol.; 1987. pp. 1–17.
- Taiz L. Annu Rev Plant Physiol. 1984;35:585–657. - PubMed
- Haughn G W, Somerville C R. Dev Genet. 1988;9:73–89.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases