Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2 - PubMed (original) (raw)
Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2
D X Tishkoff et al. Proc Natl Acad Sci U S A. 1997.
Abstract
A two-hybrid screen was used to identify Saccharomyces cerevisiae genes encoding proteins that interact with MSH2. One gene was found to encode a homologue of Schizosaccharomyces pombe EXO1, a double-stranded DNA-specific 5'-3' exonuclease. S. cerevisiae EXO1 interacted with both S. cerevisiae and human MSH2 in two-hybrid and coimmunoprecipitation experiments. exo1 mutants showed a mutator phenotype, and epistasis analysis was consistent with EXO1 functioning in the MSH2-dependent mismatch repair pathway. exo1 mutations were lethal in combination with rad27 mutations, and overexpression of EXO1 suppressed both the temperature sensitive and mutator phenotypes of rad27 mutants.
Figures
Figure 1
Interaction mating test showing target specificity of yMSH2 interactor C39. Also shown is the apparently nonspecific weak yMSH2 interactor 1-27, which expresses an activation-tagged C-terminal region of MOT1 beginning at amino acid 1579. The interaction mating test was performed as described (18). Briefly, haploid MATa strains that contain a lacZ reporter plasmid (URA3 vector) and different bait plasmids (HIS3 vector) were streaked in horizontal rows on a Ura−His− plate, and MATα strains that contain galactose-induced yMSH2 interactor plasmids (TRP1 vector) C39 and 1-27 were streaked in vertical columns on a Trp− plate. Both plates were replica plated onto a single yeast extract/peptone/dextrose (YPD) plate to allow mating, replica plated to a Ura−His−Trp− glucose plate to select diploids, then replica plated to Ura−His−Trp− 5-bromo-4-chloro-3-indolyl β-
d
-galactoside (X-Gal) glucose and galactose indicator plates and incubated for 2 days at 30°C (shown). The plasmids contained in each strain tested are indicated at the top of each column and at the right of each row.
Figure 2
Homology among the XPG/RAD2/EXO1 family of endo- and exonucleases. (A) Alignment of the N-terminal conserved regions of S. pombe EXO1, S. cerevisiae EXO1 and DIN7, and D. melanogaster Tosca. (B) Illustration of the sequence relatedness of the XPG/RAD2/EXO1 family of endo- and exonucleases adapted from figure 1 of Szankasi and Smith (7). The highly conserved N and I regions are represented by boxes filled with forward and backward diagonal lines, respectively. (C) Phylogenetic tree of the XPG/RAD2/EXO1 family of endo- and exonucleases generated from alignment of the first 240 amino acids of these proteins. YEN1, a related but divergent protein sequence found during database searches, is also included for clarity.
Figure 3
Immunoprecipitation of proteins expressed in two-hybrid strains. (A) Cell extracts (200 μg) were prepared from the two-hybrid strains expressing LexA-tagged baits and HA-tagged preys. The extracts were precleared with protein-G Sepharose, immunoprecipitated with monoclonal 12CA5 anti-HA antibody and protein-G Sepharose, fractionated by SDS/PAGE, and analyzed by Western blotting with rabbit anti-LexA antiserum. The two bands seen in every IP and PC lane are proteins nonspecifically precipitated by protein-G Sepharose. (B) To ensure appropriate expression and immunoprecipitation of HA-tagged preys, the immunoblot was stripped and reprobed with 12CA5 antibody. E, extract; IP, immunoprecipitation; PC, proteins eluted from protein G-Sepharose used to preclear.
Figure 4
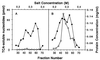
Expression of EXO1 in E. coli results in induction of an exonuclease activity. Extracts from E. coli expressing either pET28b vector alone (A) or RDK480 (B) were chromatographed on a PBE94 column. Fractions were analyzed for protein (•), exonuclease activity (○), and conductivity (NaCl, top scale). Activity is pmol acid-soluble nucleotides released per 5 μl of each fraction.
Figure 5
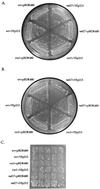
Overexpression of EXO1 suppresses both the temperature sensitive and mutator phenotypes of rad27(rth1) mutants. Isogenic wild-type (wt) (RKY2672), exo1 (RKY3044), or rad27 (RKY2608) strains were transformed with either YEp213 (a 2μ _LEU2_-marked plasmid) or pRDK480 (YEp213 with a yeast genomic insert containing the EXO1 gene) and the resulting strains streaked out on SD Leu− plates and grown for 48 h at either 30°C (A) or 37°C (B), or they were patched onto a SD Leu− plate, grown at 30°C for 2 days, then replica plated to a SD Leu− canavanine plate and grown at 30°C for 3 days to detect the presence of Canr mutations, which appear as papillae (C).
Similar articles
- Saccharomyces cerevisiae exonuclease-1 plays a role in UV resistance that is distinct from nucleotide excision repair.
Qiu J, Guan MX, Bailis AM, Shen B. Qiu J, et al. Nucleic Acids Res. 1998 Jul 1;26(13):3077-83. doi: 10.1093/nar/26.13.3077. Nucleic Acids Res. 1998. PMID: 9628902 Free PMC article. - EXO1 and MSH6 are high-copy suppressors of conditional mutations in the MSH2 mismatch repair gene of Saccharomyces cerevisiae.
Sokolsky T, Alani E. Sokolsky T, et al. Genetics. 2000 Jun;155(2):589-99. doi: 10.1093/genetics/155.2.589. Genetics. 2000. PMID: 10835383 Free PMC article. - Exonuclease 1-dependent and independent mismatch repair.
Goellner EM, Putnam CD, Kolodner RD. Goellner EM, et al. DNA Repair (Amst). 2015 Aug;32:24-32. doi: 10.1016/j.dnarep.2015.04.010. Epub 2015 Apr 30. DNA Repair (Amst). 2015. PMID: 25956862 Free PMC article. Review. - EXO1-A multi-tasking eukaryotic nuclease.
Tran PT, Erdeniz N, Symington LS, Liskay RM. Tran PT, et al. DNA Repair (Amst). 2004 Dec 2;3(12):1549-59. doi: 10.1016/j.dnarep.2004.05.015. DNA Repair (Amst). 2004. PMID: 15474417 Review.
Cited by
- Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection.
Cannavo E, Cejka P, Kowalczykowski SC. Cannavo E, et al. Proc Natl Acad Sci U S A. 2013 Apr 30;110(18):E1661-8. doi: 10.1073/pnas.1305166110. Epub 2013 Apr 15. Proc Natl Acad Sci U S A. 2013. PMID: 23589858 Free PMC article. - Sensitivity to phosphonoacetic acid: a new phenotype to probe DNA polymerase delta in Saccharomyces cerevisiae.
Li L, Murphy KM, Kanevets U, Reha-Krantz LJ. Li L, et al. Genetics. 2005 Jun;170(2):569-80. doi: 10.1534/genetics.104.040295. Epub 2005 Mar 31. Genetics. 2005. PMID: 15802517 Free PMC article. - Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication.
Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Garg P, et al. Genes Dev. 2004 Nov 15;18(22):2764-73. doi: 10.1101/gad.1252304. Epub 2004 Nov 1. Genes Dev. 2004. PMID: 15520275 Free PMC article. - EXO1 plays a role in generating type I and type II survivors in budding yeast.
Maringele L, Lydall D. Maringele L, et al. Genetics. 2004 Apr;166(4):1641-9. doi: 10.1534/genetics.166.4.1641. Genetics. 2004. PMID: 15126386 Free PMC article. - Genetic and genomic basis of the mismatch repair system involved in Lynch syndrome.
Tamura K, Kaneda M, Futagawa M, Takeshita M, Kim S, Nakama M, Kawashita N, Tatsumi-Miyajima J. Tamura K, et al. Int J Clin Oncol. 2019 Sep;24(9):999-1011. doi: 10.1007/s10147-019-01494-y. Epub 2019 Jul 4. Int J Clin Oncol. 2019. PMID: 31273487 Review.
References
- Crouse G F. In: DNA Damage and Repair. Nickoloff J A, Hoekstra M F, editors. Vol. 1. Clifton, NJ: Humana; 1997. , in press.
- Kolodner R. Genes Dev. 1996;10:1433–1442. - PubMed
- Marsischky G T, Filosi N, Kane M F, Kolodner R. Genes Dev. 1996;10:407–420. - PubMed
- Alani E, Chi N-W, Kolodner R D. Genes Dev. 1995;9:234–247. - PubMed
- Johnson R E, Gopala K K, Prakash L, Prakash S. Science. 1995;269:238–240. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 AI028691/AI/NIAID NIH HHS/United States
- AI28691/AI/NIAID NIH HHS/United States
- GM50006/GM/NIGMS NIH HHS/United States
- CA06516/CA/NCI NIH HHS/United States
- P30 CA006516/CA/NCI NIH HHS/United States
- R01 GM050006/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases