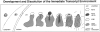Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment - PubMed (original) (raw)
Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment
A M Ishov et al. J Cell Biol. 1997.
Abstract
The development of an induced transcript environment was investigated at the supramolecular level through comparative localization of the human cytomegalovirus immediate early (IE) transcripts and specific nuclear domains shortly after infection. Compact aggregates of IE transcripts form only adjacent to nuclear domain 10 (ND10), and the viral protein IE86 accumulates exclusively juxtaposed to the subpopulation of ND10 with transcripts. The stream of transcripts is funneled from ND10 into the spliceosome assembly factor SC35 domain through the accumulation of IE86 protein, which recruits some components of the basal transcription machinery. Concomitantly the IE72 protein binds to ND10 and later disperses them. The domain containing the zinc finger region of IE72 is essential for this dispersal. Positional analysis of proteins IE86 and IE72, IE transcripts, ND10, the spliceosome assembly factor SC35, and basal transcription factors defines spatially and temporally an immediate transcript environment, the basic components of which exist in the cell before viral infection, providing the structural environment for the virus to usurp.
Figures
Figure 3
Outline of the HCMV genome depicting the IE transcript with mutations used in transfection experiments. Short lines with numbers at the upper part of the IE bar show the location of insertion mutants and brackets at the lower part show deletions in the IE1 gene.
Figure 1
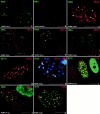
Correlation of HCMV IE gene transcripts with various nuclear domains as visualized by combined immunohistochemistry and in situ hybridization. Confocal microscopy was used to document single optical sections of WI38 cells, which are presented at high magnification. Pseudocolors were chosen to reflect either FITC labeling (green), Texas red (red), or cy5 (blue) as indicated in the upper part of each panel for the respective macromolecules or structures labeled. (a) Two infected cells showing a limited number of short and compact IE transcript-derived signals; (b) control, mock-infected cell to demonstrate probe specificity. (c) Nucleus double labeled for IE transcripts and the SC35 domain. The transcripts partially overlap with the SC35 domain. (d) In situ hybridization using the IE gene probe under denaturing conditions and after RNase treatment. Small punctate hybridization signals indicate input viral genomes, and some are found juxtaposed to ND10 (arrows). (e) Double labeling of IE transcripts and ND10. The lower cell is uninfected, and the upper cell shows the transcript signals juxtaposed to ND10. (f) Same as e but at 3 h after infection. A cell in the lower right corner shows the same configuration as at 2 h after infection, but in three nuclei seen in the center, the RNA signal is less compact, and no ND10 are seen. (g) Uninfected cell double labeled for ND10 and the SC35 domain. A large number of ND10 appear associated with the rim of the SC35 domain. (h) Infected cell triple labeled for ND10, IE transcripts, and SC35. Transcript signal appears to locate with highest concentration at ND10 and is also found in the SC35 domain. Inset shows the arrangement as expected when all three components appear in the same plane. (i) Cells double labeled for ND10 and IE72. The left nucleus is apparently not infected, the middle nucleus shows IE72 and ND10 colocalization at all ND10, and the right nucleus shows only dispersed IE72 and no ND10. (j) Cells double labeled for ND10 and IE86. The lower cell appears uninfected, the top left nucleus shows IE86 located adjacent to a few ND10, and the top right nucleus reveals IE86 throughout, with some sites of higher concentration. (k) Double labeling of IE72 and IE86. All IE86 accumulations are located next to a few IE72 sites. Bars, 10 μm.
Figure 2
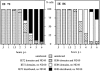
Time course of IE72 and IE86 localization relative to ND10. 500 cells were evaluated for each time point for ND10 and IE72 or ND10 and IE86. Each cell was placed in either of four categories: with ND10 only, with ND10 and either IE72 or IE86, with IE72 or IE86 in specific domains without ND10, or with only homogeneous IE72 or IE86 staining. These figures are from a single experiment with >95% infected cells at 6 h after infection (3 PFU). Minor variations in this time course were found when lower or higher PFU were used.
Figure 4
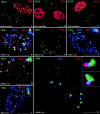
IE72, IE86, and IE transcript localization relative to other nuclear domains in human WI38 fibroblasts after transfection or infection with HCMV. (a) Cells transfected with pIE86 expressing IE86 and double labeled for ND10 and IE86. The bottom left nucleus is untransfected. The top nucleus shows a high concentration of IE86 and colocalization with ND10, which remain intact in IE86- expressing cells. (b) Cells transfected with pIE72 and double labeled for ND10 and IE72. The left transfected nucleus shows no ND10, whereas the right transfected cell is one of the few that had retained some ND10 16 h after transfection. (c) Cell transfected with dE24 expressing the putative zinc finger region deletion mutant of IE72 and double labeled for ND10 and IE72. The bottom nucleus is untransfected, and the top one shows retention of ND10 in the transfected cell. (d) HCMV-infected cell triple labeled for ND10, IE86, and the SC35 domain. IE86 domains associated with ND10 are also associated with the SC35 domain. The inset illustrates the presumed positioning if all components were in two dimensions. (e) Single nucleus double labeled for the IE86 protein and transcripts. Both signals overlap partially or appear next to each other. (f) Cell triple labeled for IE86, IE transcripts, and the SC35 domain. The IE86 cloud partly overlaps with the transcript signal, most of which is in the SC35 domain. (g) Cell triple labeled for ND10, IE transcripts, and IE86 protein. IE86 appears as a cloud surrounding the transcript exemplified in the inset. (h) Same as g but showing two cells at slightly different stages of ND10 removal. The top left nucleus still has all ND10 and only two transcript signals. The lower right nucleus has very few and smaller ND10, all of which are associated with transcript signals. The insets show one such immediate transcript environment from each nucleus. (i) HSV-1–infected cell labeled for ND10, IE1 gene transcripts, and SC35 domains, demonstrating the same general development as for HCMV. Inset shows at higher magnification that the transcript signals have the same position to ND10 and SC35 domains. Bars, 10 μm.
Figure 6
Schematic sequence of events during the immediate early steps of HCMV infection. Key to structures and shading is shown at the right.
Figure 5
Localization of the cellular proteins relative to IE86 by double labeling of fibroblasts 3 h after infection. (a–c) Separate scans of FITC signal derived from TBP (b) and IE86 (c). Superimposition (a) shows colocalization. (d–f) Separate scans of FITC signal derived from TFIIB (e) and IE86 (f). Superimposition (d) shows colocalization. (g–i) Separate scans of FITC signal derived from p53 (h) and IE86 (i). Superimposition (g) shows no enrichment of p53 at the IE86 domains. Bars, 10 μm.
Similar articles
- The human cytomegalovirus major immediate-early gene.
Stenberg RM. Stenberg RM. Intervirology. 1996;39(5-6):343-9. doi: 10.1159/000150505. Intervirology. 1996. PMID: 9130044 Review. - Activation and regulation of human cytomegalovirus early genes.
Spector DH. Spector DH. Intervirology. 1996;39(5-6):361-77. doi: 10.1159/000150507. Intervirology. 1996. PMID: 9130046 Review.
Cited by
- Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains.
Ishov AM, Vladimirova OV, Maul GG. Ishov AM, et al. J Virol. 2002 Aug;76(15):7705-12. doi: 10.1128/jvi.76.15.7705-7712.2002. J Virol. 2002. PMID: 12097584 Free PMC article. - Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections.
Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. Tavalai N, et al. J Virol. 2006 Aug;80(16):8006-18. doi: 10.1128/JVI.00743-06. J Virol. 2006. PMID: 16873257 Free PMC article. - Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression.
Negorev DG, Vladimirova OV, Ivanov A, Rauscher F 3rd, Maul GG. Negorev DG, et al. J Virol. 2006 Aug;80(16):8019-29. doi: 10.1128/JVI.02164-05. J Virol. 2006. PMID: 16873258 Free PMC article. - Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA.
Boisvert FM, Hendzel MJ, Bazett-Jones DP. Boisvert FM, et al. J Cell Biol. 2000 Jan 24;148(2):283-92. doi: 10.1083/jcb.148.2.283. J Cell Biol. 2000. PMID: 10648561 Free PMC article. - Components of nuclear domain 10 bodies regulate varicella-zoster virus replication.
Kyratsous CA, Silverstein SJ. Kyratsous CA, et al. J Virol. 2009 May;83(9):4262-74. doi: 10.1128/JVI.00021-09. Epub 2009 Feb 11. J Virol. 2009. PMID: 19211749 Free PMC article.
References
- Caswell R, Hagemeier C, Chou C-J, Hayward G, Kouzarides T, Sinclair J. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcription regulation. J Gen Virol. 1993;74:2691–2698. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources