Xeroderma pigmentosum and trichothiodystrophy are associated with different mutations in the XPD (ERCC2) repair/transcription gene - PubMed (original) (raw)
Xeroderma pigmentosum and trichothiodystrophy are associated with different mutations in the XPD (ERCC2) repair/transcription gene
E M Taylor et al. Proc Natl Acad Sci U S A. 1997.
Abstract
The xeroderma pigmentosum group D (XPD) protein has a dual function, both in nucleotide excision repair of DNA damage and in basal transcription. Mutations in the XPD gene can result in three distinct clinical phenotypes, XP, trichothiodystrophy (TTD), and XP with Cockayne syndrome. To determine if the clinical phenotypes of XP and TTD can be attributed to the sites of the mutations, we have identified the mutations in a large group of TTD and XP-D patients. Most sites of mutations differed between XP and TTD, but there are three sites at which the same mutation is found in XP and TTD patients. Since the corresponding patients were all compound heterozygotes with different mutations in the two alleles, the alleles were tested separately in a yeast complementation assay. The mutations which are found in both XP and TTD patients behaved as null alleles, suggesting that the disease phenotype was determined by the other allele. If we eliminate the null mutations, the remaining mutagenic pattern is consistent with the site of the mutation determining the phenotype.
Figures
Figure 1
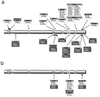
Mutations in the XPD protein. The diagrams show the XPD protein with the seven helicase domains highlighted. Amino acid changes resulting from mutations are shown boxed with the change in black on gray, the cell line designations in black on white (XP) or white on black (TTD). Subscripts 1 and 2 denote the different alleles. (a) All cell lines (mutations found in XP and TTD patients are respectively shown above and below the depicted protein). (b) Mutations found in both XP and TTD.
Figure 2
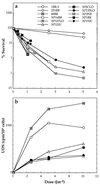
Cell survival (a) and DNA repair (UDS) (b) in fibroblast cultures of XP-D cells after UVC irradiation. 1BR.3, 251BR, and 48BR are cultures from normal donors. The rest are from XP-D donors. Results are from single experiments or means of two experiments.
Figure 3
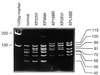
_Aci_I digests of PCR products from different cell strains. PCR product from bases 1751–2261 was digested with _Aci_I and electrophoresed on a 12.5% acrylamide gel. The C2125T or G2126A mutation results in loss of 21- and 68-bp fragments and a novel band of 89 bp (lanes 2 and 3 homozygous; lanes 4 and 5 heterozygous). The G1925C (Arg-616 → Pro) mutation causes a loss of bands at 45 and 70 bp with a novel band at 115 bp (heterozygous in lanes 5, 6, and 7).
Figure 4
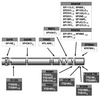
Causative mutations in the XPD protein. Designations are as in Fig. 1. Null alleles have been eliminated from Fig. 1_a_, so that only those mutations thought to be responsible for the phenotype are shown.
Similar articles
- The cancer-free phenotype in trichothiodystrophy is unrelated to its repair defect.
Berneburg M, Clingen PH, Harcourt SA, Lowe JE, Taylor EM, Green MH, Krutmann J, Arlett CF, Lehmann AR. Berneburg M, et al. Cancer Res. 2000 Jan 15;60(2):431-8. Cancer Res. 2000. PMID: 10667598 - Analysis of mutations in the XPD gene in Italian patients with trichothiodystrophy: site of mutation correlates with repair deficiency, but gene dosage appears to determine clinical severity.
Botta E, Nardo T, Broughton BC, Marinoni S, Lehmann AR, Stefanini M. Botta E, et al. Am J Hum Genet. 1998 Oct;63(4):1036-48. doi: 10.1086/302063. Am J Hum Genet. 1998. PMID: 9758621 Free PMC article. - Two individuals with features of both xeroderma pigmentosum and trichothiodystrophy highlight the complexity of the clinical outcomes of mutations in the XPD gene.
Broughton BC, Berneburg M, Fawcett H, Taylor EM, Arlett CF, Nardo T, Stefanini M, Menefee E, Price VH, Queille S, Sarasin A, Bohnert E, Krutmann J, Davidson R, Kraemer KH, Lehmann AR. Broughton BC, et al. Hum Mol Genet. 2001 Oct 15;10(22):2539-47. doi: 10.1093/hmg/10.22.2539. Hum Mol Genet. 2001. PMID: 11709541 - Molecular and cellular analysis of the DNA repair defect in a patient in xeroderma pigmentosum complementation group D who has the clinical features of xeroderma pigmentosum and Cockayne syndrome.
Broughton BC, Thompson AF, Harcourt SA, Vermeulen W, Hoeijmakers JH, Botta E, Stefanini M, King MD, Weber CA, Cole J, et al. Broughton BC, et al. Am J Hum Genet. 1995 Jan;56(1):167-74. Am J Hum Genet. 1995. PMID: 7825573 Free PMC article. Review. - Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription.
Berneburg M, Lehmann AR. Berneburg M, et al. Adv Genet. 2001;43:71-102. doi: 10.1016/s0065-2660(01)43004-5. Adv Genet. 2001. PMID: 11037299 Review.
Cited by
- MultiVERSE: a multiplex and multiplex-heterogeneous network embedding approach.
Pio-Lopez L, Valdeolivas A, Tichit L, Remy É, Baudot A. Pio-Lopez L, et al. Sci Rep. 2021 Apr 22;11(1):8794. doi: 10.1038/s41598-021-87987-1. Sci Rep. 2021. PMID: 33888761 Free PMC article. - Constructive rescue of TFIIH instability by an alternative isoform of XPD derived from a mutated XPD allele in mild but not severe XP-D/CS.
Horibata K, Kono S, Ishigami C, Zhang X, Aizawa M, Kako Y, Ishii T, Kosaki R, Saijo M, Tanaka K. Horibata K, et al. J Hum Genet. 2015 May;60(5):259-65. doi: 10.1038/jhg.2015.18. Epub 2015 Feb 26. J Hum Genet. 2015. PMID: 25716912 - The phosphorylation of the androgen receptor by TFIIH directs the ubiquitin/proteasome process.
Chymkowitch P, Le May N, Charneau P, Compe E, Egly JM. Chymkowitch P, et al. EMBO J. 2011 Feb 2;30(3):468-79. doi: 10.1038/emboj.2010.337. Epub 2010 Dec 14. EMBO J. 2011. PMID: 21157430 Free PMC article. - Cockayne syndrome and xeroderma pigmentosum.
Rapin I, Lindenbaum Y, Dickson DW, Kraemer KH, Robbins JH. Rapin I, et al. Neurology. 2000 Nov 28;55(10):1442-9. doi: 10.1212/wnl.55.10.1442. Neurology. 2000. PMID: 11185579 Free PMC article. Review.
References
- Aboussekhra A, Biggerstaff M, Shivji M K K, Vilpo J A, Moncollin V, Podust V N, Protic M, Hubscher U, Egly J-M, Wood R D. Cell. 1995;80:859–868. - PubMed
- Mu D, Park C H, Matsunaga T, Hsu D S, Reardon J T, Sancar A. J Biol Chem. 1995;270:2415–2418. - PubMed
- Wood R D. Annu Rev Biochem. 1996;65:135–167. - PubMed
- O’Donovan A, Davies A A, Moggs J G, West S C, Wood R D. Nature (London) 1994;371:432–435. - PubMed
- Sijbers A M, de Laat W L, Ariza R R, Biggerstaff M, Wei Y-F, Moggs J G, Carter K C, Shell B K, Evans E, de Jong M C, Rademakers S, de Rooij J, Jaspers N G J, Hoeijmakers J H J, Wood R D. Cell. 1996;86:811–822. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials