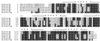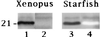Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene - PubMed (original) (raw)
Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene
S Middendorp et al. Proc Natl Acad Sci U S A. 1997.
Abstract
Among the numerous centrin isoforms identified by two-dimensional gel electrophoresis in human cells, an acidic and slow-migrating isoform is particularly enriched in a centrosome fraction. We report here that this isoform specifically reacts with antibodies raised against Saccharomyces cerevisiae Cdc31p and is present, as other centrin isoforms, in the distal lumen of centrioles. It is encoded by a new centrin gene, which we propose to name HsCEN3 (Homo sapiens centrin gene 3). This gene is more closely related to the yeast CDC31 gene, and shares less identity with algae centrin than HsCEN1 and HsCEN2. A murine CDC31-related gene was also found that shows 98% identity and 100% similarity with HsCEN3, demonstrating a higher interspecies conservation than the murine centrin gene MmCEN1 (Mus musculus centrin gene 1) with either HsCEN1, or HsCEN2. Finally, immunological data suggest that a CDC31-related gene could exist in amphibians and echinoderms as well. All together, our data suggest the existence of two divergent protein subfamilies in the current centrin family, which might be involved in distinct centrosome-associated functions. The possible implication of this new mammalian centrin gene in centrosome duplication is discussed.
Figures
Figure 1
Specificity of anti-Cdc31p Abs in human cells. (A Upper) Western blot analysis of low-speed Triton X-100 soluble (S) and insoluble (I) protein fractions and of a centrosome-enriched fraction (Ctr) obtained from human lymphoblastic KE37 cells, using the 20H5 mAb (Left) and the rabbit anti-Cdc31p Ab (Right). Both halves correspond to the same nitrocellulose filter. The Ctr lane has been split in two. (Lower) Same analysis as in A using another monoclonal anti-CrCenp Ab (11B2 mAb) and goat polyclonal anti-Cdc31p Abs. Note that the 23-kDa band recognized by anti-Cdc31p Abs corresponds to a minor band in both 20H5 mAb and mAb 11B2 signals (arrows). Proteins corresponding to 5 × 105 (Upper) and 2 × 105 (Lower) cells were loaded on each soluble and insoluble lanes. Centrosome lanes correspond to 5 × 107 and 3 × 107 centrosomes, respectively. (B) Western blot analysis of the Ctr fraction by two-dimensional electrophoresis. The same blot was first incubated with 20H5 mAb, and the signal was detected using a biotin-conjugated anti-mouse secondary Ab and alkaline phosphatase-conjugated streptavidin (Left). The blot was then incubated with the rabbit anti-Cdc31p Ab and revealed by a peroxydase-coupled anti-rabbit secondary Ab (Right). The blot is oriented with the acidic pIs on the left. Note that a single spot is identified by the anti-Cdc31p Ab, which corresponds to the isoform having the higher molecular weight and the more acidic pI in the 20H5 mAb pattern (arrows). The other spots previously stained by 20H5 mAb are seen in negative, enabling one to position accurately one pattern with respect to the other. The same result is obtained if anti-Cdc31p Abs are used first (data not shown). Proteins from 108 centrosomes were loaded. (C) Specificity of a monoclonal anti-CrCen Ab (20H5 mAb, Left) and of rabbit polyclonal anti-Cdc31p Abs (Right) on purified Cdc31p (lane 1), HsCen1p (lane 2), HsCen2p (lane 3), CrCenp (lane 4), and purified human calmodulin (lane 5). Protein (10 ng) was loaded on each lane. (D) Specificity of a monoclonal anti-CrCen Ab (20H5 mAb, Left) and of rabbit polyclonal anti-Cdc31p Abs (Right) on bacterially produced HsCen1p (lane 1), HsCen2p (lane 2), HsCen3p (lane 3), and histidine tagged-HsCen3p (lane 4). Similar amount of purified HsCen1p, HsCen2p, and of HsCen3p from a bacterial lysate (as estimated by Coomassie blue staining) were loaded on each lane, whereas a 10-fold amount of purified histidine-tagged purified HsCen3p was loaded. Arrowheads point to HsCen3p. Positions of molecular weights (31, 21, and 14 kDa) are indicated on the left-hand side of the blots.
Figure 2
Predicted protein sequences for HsCen3 and MmCen3. The two new sequences are compared with ScCdc31p and to the known centrin proteins from both human and mouse origin. Alignement was performed using the
pileup
program (Infobiogen, Villejuif, France). Comparison to ScCdc31p was performed using the
boxshade
program (Institut Suisse de Recherches Experimentales sur le Cancer, Lausanne, Switzerland). Amino acids identical to ScCdc31p and present in at least half of the sequences are boxed in black. Conservative changes are boxed in grey. The position of the four EF-hand domains is indicated by dashes (position of the helix) and stars (conserved positions of the loops or fixation pockets). The
pileup
program from Infobiogen aligned the new sequences with the known human and murine centrin sequences by creating a gap of 3 amino acids between the NH2 terminus and the conserved central part of the sequence. This suggests that a deletion or an addition in the amino terminus appeared when these genes diverged. The default parameters of the programs were used in all cases. The GenBank accession numbers for HsCen3 and MmCen3 are Y12473 and Y12474, respectively.
Figure 3
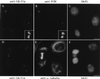
Cellular distribution of HsCen3p. Cells were triple stained with rabbit anti-Cdc31p Abs (A), an anti-PCM mAb (CTR 453, B), and 4′,6-diamidino-2-phenylindole (DAPI) (C), or with rabbit anti-Cdc31p Abs (D), an anti-α-tubulin mAb (E), and DAPI (F). A 6-fold magnification of the duplicated centrosome framed in A and B is shown on the bottom right of A and B. Note that the two distinct pairs of closely associated dots stained with anti-Cdc31p Abs correspond to one pair of duplicated centrioles. Note that the midbody joining the two sister-cells shown in D_–_F is slightly stained with the anti-Cdc31p Ab. (Bars = 10 μm.)
Figure 4
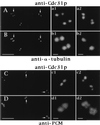
Localization of HsCen3p in isolated centrosomes. Centrosomes isolated from KE 37 cells were double-labeled with rabbit anti-Cdc31p Abs (A) and an anti-α-tubulin mAb (B) or with rabbit anti-Cdc31p Abs (C) and an anti-PCM mAb (CTR453, D). Four-fold magnification of centrosomes pointed out by arrows are shown on the right side of the figure (a1_–_b1, a2_–_b2). Given the distances between the centers of the dots obtained with anti-Cdc31p Abs, the pair figure a1_–_b1 may show two G1 centrosomes with disoriented centrioles, whereas the pair a2_–_b2 may represent a duplicating centrosome containing not fully grown daughter centrioles. Note that in this case, centrioles and their corresponding procentrioles are not resolved by the tubulin staining whereas they are clearly resolved by the anti-Cdc31p Abs staining. In C and D, the c1_–_d1 magnified pair pictures correspond to a centrosome that has not yet initiated centriole duplication according to centrin staining whereas duplication is largely engaged in the centrosome shown on c2_–_d2 pair pictures. (Bars = 5 μm in A_–_D and 0.5 μm in high magnification windows.)
Figure 5
Analysis of centrin in Xenopus and starfish eggs by Western blot analysis. Total proteins from low speed Xenopus egg extracts (Left) and from starfish fertilized eggs (Right) were submitted to SDS/PAGE on a 12% polyacrylamide gel. For Xenopus, two different lanes were incubated with 20H5 mAb (lane 1) and anti-Cdc31p Abs (lane 2), using biotin-conjugated anti-mouse and anti-rabbit secondary Abs, respectively. For starfish, the same lane was incubated first with 20H5 mAb (lane 3), and the detection performed as described, and second with rabbit anti-Cdc31p Abs (lane 4), using peroxydase-conjugated anti-rabbit secondary Abs. The slow-migrating band in the 20H5 pattern is specifically recognized by anti-Cdc31p Abs in Xenopus and starfish.
Similar articles
- Genetic interactions between CDC31 and KAR1, two genes required for duplication of the microtubule organizing center in Saccharomyces cerevisiae.
Vallen EA, Ho W, Winey M, Rose MD. Vallen EA, et al. Genetics. 1994 Jun;137(2):407-22. doi: 10.1093/genetics/137.2.407. Genetics. 1994. PMID: 8070654 Free PMC article. - Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles.
Errabolu R, Sanders MA, Salisbury JL. Errabolu R, et al. J Cell Sci. 1994 Jan;107 ( Pt 1):9-16. doi: 10.1242/jcs.107.1.9. J Cell Sci. 1994. PMID: 8175926 - Centrin/Cdc31 is a novel regulator of protein degradation.
Chen L, Madura K. Chen L, et al. Mol Cell Biol. 2008 Mar;28(5):1829-40. doi: 10.1128/MCB.01256-07. Epub 2007 Dec 26. Mol Cell Biol. 2008. PMID: 18160718 Free PMC article. - Centrosomes: Sfi1p and centrin unravel a structural riddle.
Salisbury JL. Salisbury JL. Curr Biol. 2004 Jan 6;14(1):R27-9. doi: 10.1016/j.cub.2003.12.019. Curr Biol. 2004. PMID: 14711432 Review. - A mechanistic view on the evolutionary origin for centrin-based control of centriole duplication.
Salisbury JL. Salisbury JL. J Cell Physiol. 2007 Nov;213(2):420-8. doi: 10.1002/jcp.21226. J Cell Physiol. 2007. PMID: 17694534 Review.
Cited by
- The two human centrin homologues have similar but distinct functions at Tetrahymena basal bodies.
Vonderfecht T, Cookson MW, Giddings TH Jr, Clarissa C, Winey M. Vonderfecht T, et al. Mol Biol Cell. 2012 Dec;23(24):4766-77. doi: 10.1091/mbc.E12-06-0454. Epub 2012 Oct 19. Mol Biol Cell. 2012. PMID: 23087207 Free PMC article. - Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein.
Nishi R, Okuda Y, Watanabe E, Mori T, Iwai S, Masutani C, Sugasawa K, Hanaoka F. Nishi R, et al. Mol Cell Biol. 2005 Jul;25(13):5664-74. doi: 10.1128/MCB.25.13.5664-5674.2005. Mol Cell Biol. 2005. PMID: 15964821 Free PMC article. - Defective nucleotide excision repair with normal centrosome structures and functions in the absence of all vertebrate centrins.
Dantas TJ, Wang Y, Lalor P, Dockery P, Morrison CG. Dantas TJ, et al. J Cell Biol. 2011 Apr 18;193(2):307-18. doi: 10.1083/jcb.201012093. Epub 2011 Apr 11. J Cell Biol. 2011. PMID: 21482720 Free PMC article. - The yeast centrin, cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity.
Sullivan DS, Biggins S, Rose MD. Sullivan DS, et al. J Cell Biol. 1998 Nov 2;143(3):751-65. doi: 10.1083/jcb.143.3.751. J Cell Biol. 1998. PMID: 9813095 Free PMC article. - Duplication of the Yeast Spindle Pole Body Once per Cell Cycle.
Rüthnick D, Schiebel E. Rüthnick D, et al. Mol Cell Biol. 2016 Apr 15;36(9):1324-31. doi: 10.1128/MCB.00048-16. Print 2016 May. Mol Cell Biol. 2016. PMID: 26951196 Free PMC article. Review.
References
- Paoletti A, Moudjou M, Paintrand M, Salisbury J L, Bornens M. J Cell Sci. 1996;109:3089–3102. - PubMed
- Nakayama S, Moncrief N D, Kretzinger R H. J Mol Evol. 1992;34:416–448. - PubMed
- Bhattacharya D, Steinkotter J, Melkonian M. Plant Mol Biol. 1993;23:1243–1254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases