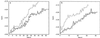Linking topography of its potential surface with the dynamics of folding of a protein model - PubMed (original) (raw)
Linking topography of its potential surface with the dynamics of folding of a protein model
R S Berry et al. Proc Natl Acad Sci U S A. 1997.
Abstract
The "3-color, 46-bead" model of a folding polypeptide is the vehicle for adapting to proteins a mode of analysis used heretofore for atomic clusters, to relate the topography of the potential surface to the dynamics that lead to formation of selected structures. The analysis is based on sequences of stationary points-successive minima, joined by saddles-that rise monotonically in energy from basin bottoms. Like structure-seeking clusters, the potential surface of the model studied here is staircase-like, rather than sawtooth-like, with highly collective motions required for passage from one minimum to the next. The surface has several deep basins whose minima correspond to very similar structures, but which are separated by high energy barriers.
Figures
Figure 1
Typical sequences of minima, with their linking saddles, rising monotonically from basin bottoms. (a) Sequences from the global minimum (primary basin). (b) Sequences from the third structure shown in the following figure (secondary basin). The numbers shown for one example in each panel are collectivity indices, the effective number of particles that move as the system passes from one local minimum to the next, along the sequences shown as solid lines.
Figure 2
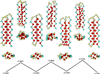
Structures and energies of the global minimum geometry and two other low-lying, basis-bottom structures of this model, and of the saddles that link them; beneath each “side” view is an “end-on” view, showing how passage between one basin bottom and another occurs by screw-like rotation and translation of one of the strands.
Figure 3
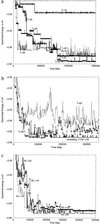
Time histories of quenched energies along typical molecular dynamics pathways taken in the folding process, showing (a) relaxation and descent when the vibrational temperature is relatively low; (b) two isothermal histories showing descent but followed by failure to remain in a single well, if the temperature is relatively high, and a contrasting example of an annealing history from approximately the same initial condition; and (c) three typical annealing histories, each leading to a different deep basin bottom. The time scale is of course scaled; if the masses of all the particles are set at m = 40 da, one time step corresponds to 1 × 10−14 s; the scale increases as .
Similar articles
- Conformational dynamics of cytochrome c: correlation to hydrogen exchange.
García AE, Hummer G. García AE, et al. Proteins. 1999 Aug 1;36(2):175-91. Proteins. 1999. PMID: 10398365 - Hydrodynamic description of protein folding: the decrease of the probability fluxes as an indicator of transition states in two-state folders.
Palyanov AY, Chekmarev SF. Palyanov AY, et al. J Biomol Struct Dyn. 2017 Nov;35(14):3152-3160. doi: 10.1080/07391102.2016.1248490. Epub 2016 Nov 7. J Biomol Struct Dyn. 2017. PMID: 27819623 - A minima hopping study of all-atom protein folding and structure prediction.
Roy S, Goedecker S, Field MJ, Penev E. Roy S, et al. J Phys Chem B. 2009 May 21;113(20):7315-21. doi: 10.1021/jp8106793. J Phys Chem B. 2009. PMID: 19391598 - Conformational dynamics and ensembles in protein folding.
Muñoz V. Muñoz V. Annu Rev Biophys Biomol Struct. 2007;36:395-412. doi: 10.1146/annurev.biophys.36.040306.132608. Annu Rev Biophys Biomol Struct. 2007. PMID: 17291180 Review. - Where the complex things are: single molecule and ensemble spectroscopic investigations of protein folding dynamics.
Takahashi S, Kamagata K, Oikawa H. Takahashi S, et al. Curr Opin Struct Biol. 2016 Feb;36:1-9. doi: 10.1016/j.sbi.2015.11.006. Epub 2015 Dec 11. Curr Opin Struct Biol. 2016. PMID: 26687767 Review.
Cited by
- Observation of strange kinetics in protein folding.
Sabelko J, Ervin J, Gruebele M. Sabelko J, et al. Proc Natl Acad Sci U S A. 1999 May 25;96(11):6031-6. doi: 10.1073/pnas.96.11.6031. Proc Natl Acad Sci U S A. 1999. PMID: 10339536 Free PMC article. - Visualizing energy landscapes with metric disconnectivity graphs.
Smeeton LC, Oakley MT, Johnston RL. Smeeton LC, et al. J Comput Chem. 2014 Jul 30;35(20):1481-90. doi: 10.1002/jcc.23643. Epub 2014 May 28. J Comput Chem. 2014. PMID: 24866379 Free PMC article. - Effects of surface tethering on protein folding mechanisms.
Friedel M, Baumketner A, Shea JE. Friedel M, et al. Proc Natl Acad Sci U S A. 2006 May 30;103(22):8396-401. doi: 10.1073/pnas.0601210103. Epub 2006 May 18. Proc Natl Acad Sci U S A. 2006. PMID: 16709672 Free PMC article. - Folding funnels and frustration in off-lattice minimalist protein landscapes.
Nymeyer H, García AE, Onuchic JN. Nymeyer H, et al. Proc Natl Acad Sci U S A. 1998 May 26;95(11):5921-8. doi: 10.1073/pnas.95.11.5921. Proc Natl Acad Sci U S A. 1998. PMID: 9600893 Free PMC article. - Topographical complexity of multidimensional energy landscapes.
Rylance GJ, Johnston RL, Matsunaga Y, Li CB, Baba A, Komatsuzaki T. Rylance GJ, et al. Proc Natl Acad Sci U S A. 2006 Dec 5;103(49):18551-5. doi: 10.1073/pnas.0608517103. Epub 2006 Nov 28. Proc Natl Acad Sci U S A. 2006. PMID: 17132739 Free PMC article.
References
- Ball K D, Berry R S, Proykova A, Kunz R E, Wales D J. Science. 1996;271:963–966.
- Kunz R E, Berry R S. J Chem Phys. 1995;103:1904–1912.
- Kunz R E, Berry R S, Astakhova T. Surf Rev Lett. 1996;3:307–312.
- Kunz, R. E., Blaudeck, P., Hoffmann, K. H. & Berry, R. S. (1997), in press.
- Vekhter B, Ball K D, Rose J, Berry R S. J Chem Phys. 1997;106:4644–4650.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources