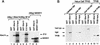Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells - PubMed (original) (raw)
Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells
M Boudjelal et al. Genes Dev. 1997.
Abstract
We report the cDNA cloning of Stra13, a novel retinoic acid (RA)-inducible gene from P19 embryonal carcinoma cells that encodes a basic helix-loop-helix (bHLH) protein that shows the highest sequence similarities to the Drosophila Hairy and Enhancer of split and mouse Hes proteins. Stra13 does not bind to the known consensus motifs (E-box and N-box) for bHLH proteins, but can repress activated transcription (through an alpha-helix rich domain) in part by interaction with general factors of the basal transcription machinery. During mouse embryogenesis, Stra13 RNA is expressed in the neuroectoderm, and also in a number of mesodermal and endodermal derivatives. Remarkably, overexpression of Stra13 in P19 cells results in neuronal differentiation in monolayer culture, under conditions where wild-type P19 cells only undergo mesodermal/endodermal differentiation. This neuronal differentiation is accompanied by an altered expression of mesodermal and neuronal markers, indicating that Stra13 could be one of the earliest RA target genes whose expression is required for repression of mesodermal/endodermal differentiation and/or induction of neuronal differentiation when P19 cell aggregates are exposed to RA. Our results raise the possibility that Stra13 could be involved as a repressor in a number of decision events occurring during differentiation of various cell lineages.
Figures
Figure 1
Nucleotide and predicted amino acid sequence of mouse Stra13 cDNA. Numbers on the right correspond to nucleotide or amino acid positions. The first and last codons of the ORF are boxed and the putative polyadenylation signal is underlined. The amino acids corresponding to the putative bHLH regions are underlined and boxed. Amino acids forming putative α-helix structures are underlined with thick lines. Glutamine residues (Q) in the glutamine-rich region and residues of the putative casein kinase 2 motif are in boldface type with a dotted underline.
Figure 2
SDS-PAGE of in vitro and in vivo synthesized Stra13 protein and induction of Stra13 expression in P19 cells by RA. (A) (Lane 1) Autoradiography of the product of Stra13 cDNA transcribed and translated in vitro and labeled with [35S] methionine. (Lanes 2,3) Immunological detection of the Stra13 protein (with polyclonal antibodies) in cytoplasmic (C, lane 2) and nuclear (N, lane 3) extracts prepared from COS cells transfected with the pSG5–Stra13 expression vector. (Lanes 4,5) Immunological detection of the endogenous Stra13 protein in nuclear extracts prepared from untreated and treated (1 μ
m
RA, 24 hr) P19 cells, respectively (the arrowhead points to a nonspecific cross-reaction). (B) Immunocytochemistry analysis. The Stra13 expression vector was transfected into COS cells and the expressed protein was revealed by immunocytochemistry with anti-Stra13 polyclonal antibodies at dilution. (C) Time course of Stra13 mRNA expression in P19 cells. P19 cells grown as monolayers were treated with ethanol or 1 μ
m
T-RA. Total RNA was extracted from RA-treated cells after 30 min, 1, 6, and 24 hr, and analyzed by RT–PCR for Stra13 RNA and the invariant 36B4 RNA (Materials and Methods). (D) Nuclear run-on transcription. Nuclei were isolated from ethanol (−RA)- or RA (+RA)-treated P19 cells. The in vitro synthesized radiolabeled RNA was hybridized to an excess of Stra13, RARβ, and 36B4 cDNAs immobilized on nylon membranes, as indicated.
Figure 3
Dimerization of Stra13 and interaction with other bHLH proteins. (A) Thirty microliters of glutathione–Sepharose preloaded with either GST (control) or GST–Stra13 protein (10 μg) were incubated with 5 μl of 35S-labeled Stra13, E12, or MASH1 (Gradwohl et al. 1996) protein translated in vitro (input lanes). Mixtures were incubated at 4°C for 2 hr in a total volume of 100 μl binding buffer (5 m
m
Tris-HCl at pH 8.0, 100 m
m
NaCl, 0.3 m
m
DTT, 10 m
m
MgCl2, 10% glycerol, 0.1% NP40), washed with the same buffer containing 250 m
m
KCl, and resolved by 12% SDS-PAGE as described in Jacq et al. (1994). 35S-Labeled Stra13 (lanes 1,2,3), E12 (lanes 4,5,6), and MASH1 (lanes 7,8,9) were revealed by autoradiography. (B) Interactions between Stra13 and general transcription factors. Glutathione–Sepharose beads preloaded as above with GST or GST–Stra13 proteins were incubated with 5 μl of 35S-labeled in vitro translated TBP (Caron et al. 1993), 10 μg of immunopurified HeLa cell TFIID complex (Brou et al. 1993), or 10 μg of recombinant TFIIB (Moncollin et al. 1992). 35S-Labeled TBP was detected by autoradiography (lanes 1,2); TBP, TAF100, and TFIIB were revealed by Western blotting (lanes 3–11) with cognate monoclonal antibodies (Jacq et al. 1994). TFIID and TFIIB input lanes contained 1 μg of protein.
Figure 4
Repression of activated transcription by Stra13. (A) Transcriptional repression activity of Stra13. One microgram of G4–ERE–tk-CAT reporter gene (see text for details) was transfected into COS cells either alone, or with the following expression vectors as indicated: GAL4 DNA-binding domain (1–147) (GAL), estrogen receptor (ER) DNA-binding domain [ER(C)], GAL–Stra13, or ER(C)–Stra13 (1 μg each); ER(HEG0), ER(C)–VP16 (Tora et al. 1989), GAL–AP2 (May et al. 1996) (200 ng each); GAL–VP16 (1 ng). When HEG0 was used, estrogen was added to the culture medium at a final concentration of 10−7
m
. Cells were harvested 48 hr after transfection, and the extracts were assayed for CAT activity that was plotted in a graph. (B) Schematic representation of deletion mutants of the GAL–Stra13 fusion protein and their corresponding repressive activities with the G4–ERE–tk–CAT reporter. The CAT activity of G4–ERE–tk–CAT upon COS cell cotransfection with vectors (1 μg) encoding the various GAL–Stra13 deletion mutants and either HEG0 or ER(C)–VP16 vector (200 ng) was expressed relative to the activity obtained with the unfused GAL4 DBD, which was taken as 100%.
Figure 5
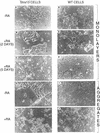
Morphological differentiation of Stra13-expressing and wild-type P19 cells cultured as monolayers or aggregates. (A–F) Phase-contrast micrography of P19 cells constitutively expressing medium levels of Stra13 (A–C) and pSG5-electroporated control cells (D–F) grown as monolayers in the absence (A,D) or after two (B,E) and 5 days culture (C,F) in the presence of 1 μ
m
T-RA. (G–J) Phase-contrast micrography of cells 5 days after plating aggregates formed by Stra13-expressing P19 cells and control cells either in the absence (G,I) or in the presence of 1 μ
m
T-RA (H,J), respectively.
Figure 6
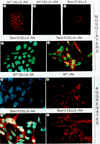
Immunofluorescence detection of SSEA-1 and βIII-tubulin expression in control and Stra13-expressing P19 cells. (A–C) Immunofluorescence detection (red color) of SSEA-1 expression in P19 control cells in the absence of T-RA (A) and after a 4 day treatment with T-RA (B), and in Stra13-expressing P19 cells cultured as monolayers in the absence of T-RA (C). (D,E) Anti-βIII-tubulin immunofluorescence (red color) of Stra13-expressing cells cultured in the absence (D) and presence (E) of T-RA for 5 days, (F–I) βIII-tubulin expression 5 days after plating cell aggregates formed by control (wild-type, F,G) and Stra13-expressing, (H,I) P19 cells untreated (F,H) or treated (G,I) with 1 μ
m
T-RA.
Figure 7
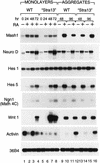
Constitutive expression of Stra13 in P19 cells results in an altered expression of neuronal and mesodermal markers. Wild-type P19 cells and “Stra13” cells were grown as monolayers or aggregates in the absence (−) or presence (+) of T-RA (100 n
m
). Total RNA was extracted from monolayers of wild-type P19 and “Stra13” cells after 24, 48, and 72 hr of RA treatment. RNA from aggregates of wild-type P19 and “Stra13” cells was isolated 2 and 4 days after plating cells. The transcripts for each gene were analyzed by semiquantitative RT–PCR, and the amount of RNA in each reaction was normalized with transcripts of the 36B4 gene (Krowczynska et al. 1989) that is unresponsive to retinoid treatment.
Figure 8
In situ analysis of Stra13 transcript expression during mouse development. Mouse embryo sections were hybridized to a 35S-labeled Stra13 antisense riboprobe; all panels are composed of a bright-field view (left) and a dark-field view (right) of the same section, where signal grain appears as white dots. (A) Sagittal section of a 9.5 dpc embryo. (B,C) Two serial coronal sections through the forebrain, heart, and forelimbs of an 11.5 dpc embryo. The section in B crosses one of the eyes, as well as the heart outflow tract. (D) Transverse section of the cervical spinal cord of a 12.5 dpc embryo. The open arrow indicates enhanced labeling in the ventral region of the ventricular layer and the filled arrow points to the signal in the ventral horn. (E) Coronal section of the head of a 15.5 dpc fetus. (F) Sagittal section at the level of the lower incisor bud of a 17.5 dpc fetus. (G,H) Two coronal sections of a 13.5 dpc fetus. G is at the level of the liver and the apex of the heart ventricles. H is a more dorsal section crossing the lungs, rib cage, and scapula. (I) Sagittal section through the stomach and kidney of a 17.5 dpc fetus. (J) Mid-sagittal section through the rectum, urinary bladder, and urethra of a 17.5 dpc fetus. Abbreviations: (ab) alveolar bone; (ao) aorta; (b) urinary bladder; (D) dorsal; (ey) eye; (fb) forebrain; (fl) forelimb; (hb) hindbrain; (hd) head; (ht) heart; (ib) incisor bud; (in) intestine; (ki) kidney; (le) lens; (li) liver; (lu) lung; (m) muscle; (mb) midbrain; (ml) mantle layer; (ne) nasal epithelium; (ng) nasal gland; (oe) oesophagus; (os) optic stalk; (ot) outflow tract of the heart; (pr) prospective retinal pigment epithelium; (pv) prevertebra; (r) radius; (r5) rhombomere 5; (re) rectum; (ri) rib; (sc) spinal cord; (sp) scapula; (st) stomach; (to) tongue; (u) ulna; (ur) urethra; (V) ventral; (vl) ventricular layer.
Similar articles
- Neurogenin1 is sufficient to induce neuronal differentiation of embryonal carcinoma P19 cells in the absence of retinoic acid.
Kim S, Yoon YS, Kim JW, Jung M, Kim SU, Lee YD, Suh-Kim H. Kim S, et al. Cell Mol Neurobiol. 2004 Jun;24(3):343-56. doi: 10.1023/b:cemn.0000022767.74774.38. Cell Mol Neurobiol. 2004. PMID: 15206818 - mSharp-1/DEC2, a basic helix-loop-helix protein functions as a transcriptional repressor of E box activity and Stra13 expression.
Azmi S, Sun H, Ozog A, Taneja R. Azmi S, et al. J Biol Chem. 2003 May 30;278(22):20098-109. doi: 10.1074/jbc.M210427200. Epub 2003 Mar 25. J Biol Chem. 2003. PMID: 12657651 - Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms.
Sun H, Taneja R. Sun H, et al. Proc Natl Acad Sci U S A. 2000 Apr 11;97(8):4058-63. doi: 10.1073/pnas.070526297. Proc Natl Acad Sci U S A. 2000. PMID: 10737769 Free PMC article. - [Expression and Regulation of Tal2 during Neuronal Differentiation in P19 Cells].
Kobayashi T. Kobayashi T. Yakugaku Zasshi. 2017;137(1):61-71. doi: 10.1248/yakushi.16-00176. Yakugaku Zasshi. 2017. PMID: 28049897 Review. Japanese.
Cited by
- BHLHE41, a transcriptional repressor involved in physiological processes and tumor development.
Bret C, Desmots-Loyer F, Moreaux J, Fest T. Bret C, et al. Cell Oncol (Dordr). 2024 Sep 10. doi: 10.1007/s13402-024-00973-3. Online ahead of print. Cell Oncol (Dordr). 2024. PMID: 39254779 Review. - Bhlhe40 Regulates Proliferation and Angiogenesis in Mouse Embryoid Bodies under Hypoxia.
Acosta-Iborra B, Gil-Acero AI, Sanz-Gómez M, Berrouayel Y, Puente-Santamaría L, Alieva M, Del Peso L, Jiménez B. Acosta-Iborra B, et al. Int J Mol Sci. 2024 Jul 12;25(14):7669. doi: 10.3390/ijms25147669. Int J Mol Sci. 2024. PMID: 39062912 Free PMC article. - Vitamin a potentiates sheep myoblasts myogenic differentiation through BHLHE40-modulated ID3 expression.
Song P, Zhao J, Zhang W, Li X, Ji B, Zhao J. Song P, et al. BMC Genomics. 2024 Mar 5;25(1):244. doi: 10.1186/s12864-024-10161-0. BMC Genomics. 2024. PMID: 38443816 Free PMC article. - GSDMD gene knockout alleviates hyperoxia-induced hippocampal brain injury in neonatal mice.
Challa NVD, Chen S, Yuan H, Duncan MR, Moreno WJ, Bramlett H, Dietrich WD, Benny M, Schmidt AF, Young K, Wu S. Challa NVD, et al. J Neuroinflammation. 2023 Sep 7;20(1):205. doi: 10.1186/s12974-023-02878-8. J Neuroinflammation. 2023. PMID: 37679766 Free PMC article. - Role of TET1-mediated epigenetic modulation in Alzheimer's disease.
Armstrong MJ, Jin Y, Vattathil SM, Huang Y, Schroeder JP, Bennet DA, Qin ZS, Wingo TS, Jin P. Armstrong MJ, et al. Neurobiol Dis. 2023 Sep;185:106257. doi: 10.1016/j.nbd.2023.106257. Epub 2023 Aug 8. Neurobiol Dis. 2023. PMID: 37562656 Free PMC article.
References
- Akazawa C, Sasai Y, Nakanishi S, Kagemaya R. Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J Biol Chem. 1992;267:21879–21885. - PubMed
- Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72:233–245. - PubMed
- Anthony-Cahill SJ, Benfield PA, Fairman R, Wasserman ZR, Brenner SL, Stafford WF, Altenbach C, Hubbell WL, de Grado WF. Molecular characterization of helix-loop-helix peptides. Science. 1992;255:979–983. - PubMed
- Bain G, Ray WJ, Yao M, Gottlieb DI. From embryonal carcinoma cells to neurons: The P19 pathway. BioEssays. 1994;16:343–348. - PubMed
- Banerjee A, Roach MC, Trcka P, Luduena RF. Increased microtubule assembly in bovine brain tubulin lacking the type III isotype of β-tubulin. J Biol Chem. 1990;265:1794–1799. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases