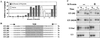Selection and nuclear immobilization of exportable RNAs - PubMed (original) (raw)
Selection and nuclear immobilization of exportable RNAs
C Grimm et al. Proc Natl Acad Sci U S A. 1997.
Abstract
The intracellular distribution of RNAs depends on interactions of cis-acting nuclear export elements or nuclear retention elements with trans-acting nuclear transport or retention factors. To learn about the relationship between export and retention, we isolated RNAs that are exported from nuclei of Xenopus laevis oocytes even when most RNA export is blocked by an inhibitor of Ran-dependent nucleocytoplasmic transport, the Matrix protein of vesicular stomatitis virus. Export of the selected RNAs is saturable and specific. When present in chimeric RNAs, the selected sequences acted like nuclear export elements in promoting efficient export of RNAs that otherwise are not exported; the pathway used for export of these chimeric RNAs is that used for the selected RNAs alone. However, these chimeric RNAs, unlike the selected RNAs, were not exported in the presence of Matrix protein; thus, the nonselected sequences can cause retention of the selected RNA sequences under conditions of impaired nucleocytoplasmic transport. We propose that most RNAs are transiently immobilized in the nucleus and that release of these RNAs is an essential and early step in export. Release correlates with functional Ran-dependent transport, and the lack of export of chimeric RNAs may result from interference with the Ran system.
Figures
Figure 1
Selection of RNAs containing NEEs. (A) Enrichment of injected RNA pools for RNAs containing NEEs. The percentage of exported RNAs at 2 hr (rounds 1–12 and 17 and 18) or 24 hr (rounds 13–16) after nuclear injection was calculated as [C/(N+C)] × 100. Rounds of selection are indicated at the bottom. Solid bars, selection in presence of M protein; open bars, counterselection on ice. (B) Sequences of ET-RNAs (
E
xceptional
T
ransport RNAs) after 18 rounds of selection. Shaded box represents the randomized region (N20). Δ indicates nucleotides that were deleted from the fixed sequence during the selection procedure and arrows indicate the 3′ ends of the primers used for reverse transcription and PCR. (C) Export of the monomeric (ET-202) and dimeric (ET-202/di) forms of the winner RNA ET-202 (Top). Both forms of ET-202 RNA were uncapped, whereas the control RNAs (U1Sm− and U3; Bottom) were m7GpppG capped. RNAs were injected into nuclei of oocytes that did (+) or did not (−) contain M protein and prepared from nuclei (N) and cytoplasms (C) after 1 and 3 hr. Nucleocytoplasmic distribution of the RNAs was determined by denaturing 8% PAGE containing 7 M urea. I, input RNA.
Figure 5
Nuclear retention of chimeric RNAs. (A) The ability of the selected NEEs in chimeric RNAs to support export in the absence or presence of M protein or mAb414 was tested by injection of the RNAs into nuclei of control oocytes (−) or oocytes containing M protein or mAb414. Export of chimeric U1124/ET-202 (Top) and U1124/ET-208 (Upper Middle) RNAs was tested in oocytes containing M protein or mAb414, as indicated; export of U1124 RNA and ET-202 RNA is shown for comparison. All RNAs were ApppG capped (except for ET-202 RNA, which was uncapped). RNAs were isolated from nuclear (N) and cytoplasmic (C) fractions 1 and 3 hr after nuclear injection and analyzed as in Fig. 1_C_. (B) Specificity of export and retention of the chimeric RNAs was tested by coinjection of a mixture of unlabeled RNAs containing 1.0 pmol of ApppG-capped U2 RNA and 0.5 pmol of NL-15 RNA; the former RNA served as competitor for nonselected RNA sequences present in the chimeric ET-202/U2Sm− RNA and the latter RNA was a competitor for nonspecific binding to nuclear La protein. Intracellular distribution of the chimeric RNA was assayed as in Fig. 5_A_.
Figure 2
Intracellular distributions of sequence variants of ET-202 RNAs. Mutated templates of ET-202, generated by error-prone PCR, were transcribed to make a pool of variant ET-202 RNAs that were injected into nuclei of oocytes containing M protein. RNAs isolated from the cytoplasm (C) or nucleus (N) after 2 hr of incubation were reverse transcribed, cloned, and sequenced. Underlined nucleotides in the ET-202 sequence represent the ends of the primers used in the PCR amplification. ∗ indicates individual RNAs that were tested further for their abilities to be exported to the presence of M protein. Nucleotides are shown that differ from the original ET-202 RNA; an outlined letter represents inserted nucleotides and Δ represents a deleted nucleotide. The average numbers of changes in the RNAs were 2.4 and 5.5 per molecule in the C and N clones, respectively.
Figure 3
Common factors among exported RNAs. Competition for export of other RNAs by ET-202 RNA. One to 10 fmol of 32P-labeled ET-202 (Top), ET-208 RNA (Middle), or hY1 RNA (Bottom) were injected into nuclei of control oocytes in the absence (−) or presence of 0.5 pmol unlabeled competitor RNAs as indicated across the top, except 5.0 pmol of tRNA were used in the right-most panel; the preparation of labeled hY1 RNA also contained 0.5 pmol NL-15 RNA, to bind the nuclear La protein, thereby allowing efficient export of the hY1 RNA (5). Most RNAs were prepared from nuclear (N) and cytoplasmic (C) fractions 1, 2, and 4 hr after nuclear injection, but samples containing hY1 RNA (either labeled or as competitor) were isolated at 0.5, 1, 2, and 6 hr and samples with tRNA were isolated at 0.5, 2, and 4 hr. All RNAs were uncapped and were analyzed as in Fig. 1_C_.
Figure 4
Transposable NEEs in the selected RNAs. ET-202 RNA in the sense (a, b, e, f, i, and l) or antisense (m) orientation, SLX RNA (unselected RNAs from round 2; c and g), or ET-208 RNA (j) were fused to the 5′ or 3′ end of U6 RNA (a_–_c), U2Sm− RNA (e_–_g), U1124 RNA (i and j), or AdML pre-mRNA (l and m) as indicated. Export of these chimeric RNAs and the control RNAs U6Xho (d), U2Sm− (h), U1124 (k), and AdML pre-mRNA (n) was tested in control oocytes. Export of intron-containing AdML pre-mRNAs (l_–_n) was analyzed in the presence of 300 fmol of unlabeled, m7G-capped AdML pre-mRNA serving as competitor for splicing. RNAs containing U6 snRNA (a_–_d) were γpppG capped, whereas all other labeled RNAs (e_–_n) were ApppG capped. RNAs were isolated from nuclear (N) and cytoplasmic (C) fractions 1 and 3 hr (a_–_k) or 1, 3, and 5.5 hr (l_–_m) after nuclear injection and analyzed as in Fig. 1_C_.
Similar articles
- Identification of a novel cis-acting RNA element involved in nuclear export of hY RNAs.
Rutjes SA, Lund E, van der Heijden A, Grimm C, van Venrooij WJ, Pruijn GJ. Rutjes SA, et al. RNA. 2001 May;7(5):741-52. doi: 10.1017/s1355838201002503. RNA. 2001. PMID: 11350038 Free PMC article. - Rev-mediated nuclear export of RNA is dominant over nuclear retention and is coupled to the Ran-GTPase cycle.
Fischer U, Pollard VW, Lührmann R, Teufel M, Michael MW, Dreyfuss G, Malim MH. Fischer U, et al. Nucleic Acids Res. 1999 Nov 1;27(21):4128-34. doi: 10.1093/nar/27.21.4128. Nucleic Acids Res. 1999. PMID: 10518602 Free PMC article. - In vivo selection of RNAs that localize in the nucleus.
Grimm C, Lund E, Dahlberg JE. Grimm C, et al. EMBO J. 1997 Feb 17;16(4):793-806. doi: 10.1093/emboj/16.4.793. EMBO J. 1997. PMID: 9049308 Free PMC article. - Use of intact Xenopus oocytes in nucleocytoplasmic transport studies.
Panté N. Panté N. Methods Mol Biol. 2006;322:301-14. doi: 10.1007/978-1-59745-000-3_21. Methods Mol Biol. 2006. PMID: 16739732 Review. - Nuclear transport of RNAs in microinjected Xenopus oocytes.
Terns MP, Goldfarb DS. Terns MP, et al. Methods Cell Biol. 1998;53:559-89. doi: 10.1016/s0091-679x(08)60895-x. Methods Cell Biol. 1998. PMID: 9348525 Review. No abstract available.
Cited by
- At the Intersection of Biomaterials and Gene Therapy: Progress in Non-viral Delivery of Nucleic Acids.
Uludag H, Ubeda A, Ansari A. Uludag H, et al. Front Bioeng Biotechnol. 2019 Jun 4;7:131. doi: 10.3389/fbioe.2019.00131. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 31214586 Free PMC article. Review. - Multiple vesiculoviral matrix proteins inhibit both nuclear export and import.
Petersen JM, Her LS, Dahlberg JE. Petersen JM, et al. Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8590-5. doi: 10.1073/pnas.151240998. Epub 2001 Jul 10. Proc Natl Acad Sci U S A. 2001. PMID: 11447272 Free PMC article. - The human homologue of Saccharomyces cerevisiae Gle1p is required for poly(A)+ RNA export.
Watkins JL, Murphy R, Emtage JL, Wente SR. Watkins JL, et al. Proc Natl Acad Sci U S A. 1998 Jun 9;95(12):6779-84. doi: 10.1073/pnas.95.12.6779. Proc Natl Acad Sci U S A. 1998. PMID: 9618489 Free PMC article. - Identification of a novel cis-acting RNA element involved in nuclear export of hY RNAs.
Rutjes SA, Lund E, van der Heijden A, Grimm C, van Venrooij WJ, Pruijn GJ. Rutjes SA, et al. RNA. 2001 May;7(5):741-52. doi: 10.1017/s1355838201002503. RNA. 2001. PMID: 11350038 Free PMC article. - The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway.
Pasquinelli AE, Ernst RK, Lund E, Grimm C, Zapp ML, Rekosh D, Hammarskjöld ML, Dahlberg JE. Pasquinelli AE, et al. EMBO J. 1997 Dec 15;16(24):7500-10. doi: 10.1093/emboj/16.24.7500. EMBO J. 1997. PMID: 9405378 Free PMC article.
References
- Chang D D, Sharp P A. Cell. 1989;59:789–795. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous