A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus - PubMed (original) (raw)
A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus
M Brannon et al. Genes Dev. 1997.
Abstract
The Wnt pathway regulates the early dorsal-ventral axis in Xenopus through a complex of beta-catenin and HMG box transcription factors of the Lef/Tcf family. We show that the promoter of the dorsalizing homeo box gene siamois is a direct target for the beta-catenin/XTcf-3 complex, establishing a link between the Wnt pathway and the activation of genes involved in specifying the dorsal axis. By injecting siamois reporter constructs into the animal pole of Xenopus embryos, we show that a 0.8-kb fragment of the siamois promoter is strongly activated by beta-catenin. The proximal 0.5 kb, which is also activated by beta-catenin, contains three Lef/Tcf-binding sites. Mutations in these sites eliminate the beta-catenin-mediated activation of siamois and show that siamois is regulated by the beta-catenin/XTcf-3 complex, in combination with additional transcriptional activators. When expressed at the equator of the embryo, the siamois promoter is activated to much higher levels on the dorsal side than the ventral side. Ectopic ventral expression of beta-catenin raises the ventral expression of the siamois promoter to the dorsal levels. Conversely, ectopic dorsal expression of dominant-negative XTcf-3 abolishes the dorsal activation of the siamois promoter. Furthermore, elimination of the Lef/Tcf sites elevates the ventral expression of siamois, revealing a repressive role for XTcf-3 in the absence of beta-catenin. Finally, we find that the endogenous siamois activator, although present throughout the dorsal side of the embryo, is most potent in the dorsal vegetal region. We propose that the dorsal activation of siamois by the beta-catenin/XTcf-3 complex combined with the ventral repression of siamois by XTcf-3 results in the restriction of endogenous siamois expression to the dorsal side of Xenopus embryos.
Figures
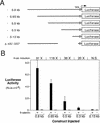
Figure 1
β-Catenin responsive regions of the siamois promoter. (A) Diagram of the siamois promoter deletion constructs used in this study. The length that each promoter fragment extends upstream of the siamois translation start site is indicated at left; each horizontal line represents the relative length of the promoter fragments. Δ−45/−357 represents a 312-bp deletion of the siamois promoter proximal to the TATA box, which is represented by an open box. Each siamois promoter fragment was fused to a luciferase reporter gene at the same point, as indicated. (B) β-Catenin responsive regions of the siamois promoter. The indicated constructs were injected into the animal pole of both blastomeres of two-cell embryos in the presence (+) or absence (−) of β-catenin RNA. Three pools of five stage 10 Xenopus embryos were assayed, and the mean and standard error of the resulting luciferase activities, in relative luciferase units (RLUs), are shown. The average fold induction by β-catenin (β-cat. Induction) is indicated above each data set. (N.S.) No significant β-catenin induction.
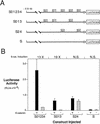
Figure 2
Determination of siamois promoter regions responsive to a dorsally localized endogenous activity. (A) Regions of the siamois promoter responsive to the dorsalizing activity. The indicated constructs were injected into the marginal zone of dorsal (D) or ventral (V) blastomeres at the four-cell stage. The mean and standard error, in RLUs, from three pools of five embryos each are shown. The average fold induction (D/V Ratio) for each construct is indicated above each data set. (N.S.) No significant dorsal vs. ventral difference. (B) siamois activation by the endogenous dorsalizing activity requires the proximal promoter region. The 0.8-kb and Δ−45/−357 reporter constructs were assayed as described above. The siamois promoter is no longer responsive to the endogenous dorsalizing activity when the proximal region containing the three Lef/Tcf sites is deleted.
Figure 3
Nucleotide sequence of the Xenopus siamois promoter −0.8-kb fragment. The first 23 nucleotides of the siamois coding sequence with its deduced amino acids and 817 nucleotides of 5′ siamois promoter sequence are shown. The transcription start site is indicated with an arrow and the TATA box is underlined. The possible Lef/Tcf-binding sites are boxed and identified above the sequence, as S0, S1, S2, S3 and S4. S0, S1, and S3 conform to the Lef/Tcf-binding site consensus, whereas S2 and S4 diverge at the 3′-most base. Nucleotides mutated in the Lef/Tcf-binding sites are underlined. The GenBank accession no. for the Xenopus siamois promoter is AF016226.
Figure 4
Lef-1 and XTcf-3 specifically bind to the siamois promoter. (A) Lef-1 binding to the siamois promoter is competed by an oligonucleotide containing a consensus Lef/Tcf-binding site. Double-stranded competitor oligonucleotides containing either a consensus Lef/Tcf-binding site (WT) or a mutated site were incubated with Lef-1 in the presence of the siamois promoter probe S1234, which contains four potential binding sites. Binding to the promoter was analyzed by electrophoretic mobility shift assays. The concentrations of competitor oligonucleotide are indicated. [(Ctr) control; uncharged reticulocyte lysate]. (B) Lef-1 and XTcf-3 form a ternary complex with β-catenin on the siamois promoter. Control lysate or lysate containing either Lef-1, ΔNLef-1, XTcf-3, or ΔNXTcf-3 were incubated with the siamois promoter probe S1234 in the presence or absence of recombinant β-catenin. β-Catenin decreases the mobility of the S1234 probe in the presence of full-length Lef-1 or XTcf-3 but not in the presence of amino-terminally truncated proteins (cf. lanes 4 and 8 with lanes 6 and 10). A nonspecific band is observed in lanes containing recombinant β-catenin. (C) Lef/Tcf consensus S1 and S3 are required for Lef-1 to bind the siamois promoter. Control reticulocyte lysate (lanes 1–4) or lysate containing Lef-1 (lanes 5–8) was incubated with the following siamois promoter probes. Probe S1234 is wild-type for all sites, S13 contains the consensus Lef/Tcf-binding S1 and S3, S24 contains the imperfect S2 and S4, and S contains none of these sites. (D) Lef-1 binds to both consensus Lef/Tcf sites S1 and S3 and causes DNA bending. Control lysate or lysate containing Lef-1 was incubated with siamois promoter probes S1234, S234, or S124, as indicated. Because sites S2 and S4 do not bind Lef-1 (as shown in Fig. 4C), binding of Lef-1 to either S1 or S3 independently is being analyzed. Note the different migration rates of probes S234 (lane 7) and S124 (lane 8) in the presence of Lef-1, which is consistent with Lef-1 DNA bending.
Figure 6
Lef/Tcf site mutations eliminate the β-catenin response of the siamois promoter. (A) Schematic representation of the constructs that result from introducing site-directed mutations into the −0.8-kb siamois promoter. S01234 contains no mutations, S013 is mutant at S2 and S4, S24 is mutant at S0, S1, and S3, and S is mutant at all sites. (B) Lef/Tcf consensus sites S0, S1, and S3 are β-catenin response elements, whereas S2 and S4 have a general activating function. The indicated promoter constructs were injected into the animal pole of both blastomeres of two-cell-stage embryos in the presence (+) or absence (−) of β-catenin. The mean luciferase activities and standard errors, in RLUs, from three pools of five embryos each are shown. Average fold inductions by β-catenin (β-cat. Induction) are indicated above each data set. Note that basal luciferase levels resulting from injection of S24 are greater than those for S01234. (N.S.) No significant β-catenin induction.
Figure 5
DNase I footprint analysis of the S1234 siamois promoter fragment. S1234 was 3′-end labeled and incubated in the presence of 0.1 μg BSA (−, lane 2), or 0.025 μg (lane 3), 0.05 μg (lane 4), or 0.1 μg (lane 5) of Lef-1 HMG domain protein. (Lane 1) Maxam-Gilbert G + A sequence reaction of the same DNA fragment. The positions and sequences of S1, S2, and S3 are indicated. DNase I-protected regions can be observed at S1 and S3 in the presence of 0.1 μg Lef-1 HMG protein (cf. lanes 2 and 5) but not S2 (shown) or S4 (not shown).
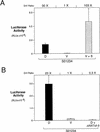
Figure 7
siamois promoter activation is dependent on the β-catenin/XTcf-3 complex. (A) β-Catenin activates the siamois promoter in the ventral marginal zone. Construct S01234 was injected dorsally (D), ventrally (V), or ventrally in the presence of β-catenin RNA (V + β). The mean and standard errors of luciferase activities are shown for each sample, as described previously. Average fold induction (D/V Ratio) for D vs. V and V + β vs. V is shown above the graph. (B) ΔNXTcf-3 blocks the dorsal activation of the siamois promoter. Construct S01234 was injected dorsally (D), ventrally (V), or dorsally in the presence of ΔNXTcf-3 RNA (D + ΔNXTcf-3). The mean and standard errors of luciferase activities are shown for each sample. Average fold inductions for D vs. V and D + ΔNXTcf-3 vs. V are shown above the graph.
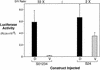
Figure 8
The Lef/Tcf-binding sites repress ventral siamois activation. Constructs S01234 and S24, which lacks the three Lef/Tcf-binding S0, S1, and S3, were injected into the marginal zone of dorsal (D) or ventral (V) blastomeres at the four-cell stage. The mean and standard errors of the resulting luciferase activities from three pools of five embryos each are shown. Average fold induction (D/V Ratio) for D vs. V is shown above both data sets. Note that construct S24 has 20-fold higher ventral activation of the siamois promoter than S01234, indicating that the siamois promoter lacking the Lef/Tcf-binding sites is no longer repressed on the ventral side of the embryo.
Figure 9
Localization of the endogenous dorsalizing activity in the embryo. (A) Diagram of a 32-cell stage Xenopus embryo showing the four tiers of blastomeres. (An) Animal pole; (Veg) vegetal pole. (B) The endogenous dorsalizing activity is highest in dorsal vegetal cells. A single dorsal or ventral blastomere at the 32-cell stage was injected with S01234. The luciferase activity of a pool of five embryos injected into the same blastomere indicates that the endogenous dorsalizing activity is present mainly in the dorsal C and D tiers (dorsal vegetal cells) of early embryos. This region of the embryo is most abundant in β-catenin (Larabell et al. 1997).
Similar articles
- VegT activation of the early zygotic gene Xnr5 requires lifting of Tcf-mediated repression in the Xenopus blastula.
Hilton E, Rex M, Old R. Hilton E, et al. Mech Dev. 2003 Oct;120(10):1127-38. doi: 10.1016/j.mod.2003.08.004. Mech Dev. 2003. PMID: 14568102 - Functional diversity of Xenopus lymphoid enhancer factor/T-cell factor transcription factors relies on combinations of activating and repressing elements.
Gradl D, König A, Wedlich D. Gradl D, et al. J Biol Chem. 2002 Apr 19;277(16):14159-71. doi: 10.1074/jbc.M107055200. Epub 2002 Jan 30. J Biol Chem. 2002. PMID: 11821382 - The Yin-Yang of TCF/beta-catenin signaling.
Barker N, Morin PJ, Clevers H. Barker N, et al. Adv Cancer Res. 2000;77:1-24. doi: 10.1016/s0065-230x(08)60783-6. Adv Cancer Res. 2000. PMID: 10549354 Review. - From cortical rotation to organizer gene expression: toward a molecular explanation of axis specification in Xenopus.
Moon RT, Kimelman D. Moon RT, et al. Bioessays. 1998 Jul;20(7):536-45. doi: 10.1002/(SICI)1521-1878(199807)20:7<536::AID-BIES4>3.0.CO;2-I. Bioessays. 1998. PMID: 9723002 Review.
Cited by
- Function of Wnt/β-catenin in counteracting Tcf3 repression through the Tcf3-β-catenin interaction.
Wu CI, Hoffman JA, Shy BR, Ford EM, Fuchs E, Nguyen H, Merrill BJ. Wu CI, et al. Development. 2012 Jun;139(12):2118-29. doi: 10.1242/dev.076067. Epub 2012 May 9. Development. 2012. PMID: 22573616 Free PMC article. - Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors.
Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Maretto S, et al. Proc Natl Acad Sci U S A. 2003 Mar 18;100(6):3299-304. doi: 10.1073/pnas.0434590100. Epub 2003 Mar 7. Proc Natl Acad Sci U S A. 2003. PMID: 12626757 Free PMC article. - Alternative splicing of Tcf7l2 transcripts generates protein variants with differential promoter-binding and transcriptional activation properties at Wnt/beta-catenin targets.
Weise A, Bruser K, Elfert S, Wallmen B, Wittel Y, Wöhrle S, Hecht A. Weise A, et al. Nucleic Acids Res. 2010 Apr;38(6):1964-81. doi: 10.1093/nar/gkp1197. Epub 2009 Dec 30. Nucleic Acids Res. 2010. PMID: 20044351 Free PMC article. - Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal.
Pereira L, Yi F, Merrill BJ. Pereira L, et al. Mol Cell Biol. 2006 Oct;26(20):7479-91. doi: 10.1128/MCB.00368-06. Epub 2006 Aug 7. Mol Cell Biol. 2006. PMID: 16894029 Free PMC article. - Interaction among GSK-3, GBP, axin, and APC in Xenopus axis specification.
Farr GH 3rd, Ferkey DM, Yost C, Pierce SB, Weaver C, Kimelman D. Farr GH 3rd, et al. J Cell Biol. 2000 Feb 21;148(4):691-702. doi: 10.1083/jcb.148.4.691. J Cell Biol. 2000. PMID: 10684251 Free PMC article.
References
- Bauer DV, Moody SA. The cleavage stage origin of Spemann’s organizer: Analysis of the movements of blastomere clones before and during gastrulation in Xenopus. Development. 1994;120:1179–1189. - PubMed
- Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. - PubMed
- Brannon M, Kimelman D. Activation of Siamois by the Wnt pathway. Dev Biol. 1996;180:344–347. - PubMed
- Brunner E, Peter O, Schweizer L, Basler K. pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature. 1997;385:829–833. - PubMed
- Carnac G, Kodjabachian L, Gurdon JB, Lemaire P. The homeobox gene Siamois is a target of the Wnt dorsalization pathway and triggers organiser activity in the absence of mesoderm. Development. 1996;122:3055–3065. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 HD007183/HD/NICHD NIH HHS/United States
- HD27262/HD/NICHD NIH HHS/United States
- F32 HD008016/HD/NICHD NIH HHS/United States
- R01 HD027262/HD/NICHD NIH HHS/United States
- 5T32HDO7183-17/HD/NICHD NIH HHS/United States
- HD08016/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials