Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family - PubMed (original) (raw)
Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family
D Reczek et al. J Cell Biol. 1997.
Abstract
Members of the ezrin-radixin-moesin (ERM) family of membrane-cytoskeletal linking proteins have NH2- and COOH-terminal domains that associate with the plasma membrane and the actin cytoskeleton, respectively. To search for ERM binding partners potentially involved in membrane association, tissue lysates were subjected to affinity chromatography on the immobilized NH2-terminal domains of ezrin and moesin, which comprise the ezrin-radixin-moesin-association domain (N-ERMAD). A collection of polypeptides at 50-53 kD from human placenta and at 58-59 kD from bovine brain bound directly to both N-ERMADs. The 50-53-kD placental proteins migrated as a major 50-kD species after phosphatase treatment, indicating that the heterogeneity is due to different phosphorylation states. We refer to these polypeptides as ERM-binding phosphoprotein 50 (EBP50). Sequence analysis of human EBP50 was used to identify an approximately 2-kb human cDNA that encodes a 357-residue polypeptide. Recombinant EBP50 binds tightly to the N-ERMADs of ezrin and moesin. Peptide sequences from the brain candidate indicated that it is closely related to EBP50. EBP50 has two PSD-95/DlgA/ZO-1-like (PDZ) domains and is most likely a homologue of rabbit protein cofactor, which is involved in the protein kinase A regulation of the renal brush border Na+/H+ exchanger. EBP50 is widely distributed in tissues, and is particularly enriched in those containing polarized epithelia. Immunofluorescence microscopy of cultured cells and tissues revealed that EBP50 colocalizes with actin and ezrin in the apical microvilli of epithelial cells, and immunoelectron microscopy demonstrated that it is specifically associated with the microvilli of the placental syncytiotrophoblast. Moreover, EBP50 and ezrin can be coimmunoprecipitated as a complex from isolated human placental microvilli. These findings show that EBP50 is a physiologically relevant ezrin binding protein. Since PDZ domains are known to mediate associations with integral membrane proteins, one mode of membrane attachment of ezrin is likely to be mediated through EBP50.
Figures
Figure 4
Nucleotide and derived protein sequence of human EBP50 cDNA. Residues matching the two placental γ-EBP50 peptide sequences are underlined. These sequence data are available from EMBL/GenBank/DDBJ under accession number AF015926.
Figure 5
Human EBP50 has homology to rabbit protein cofactor and human TKA-1. Identities are boxed. Sequences corresponding to the ∼90-residue PDZ domains in each of these proteins are underlined with thick lines. The locations of sequences of the three peptides derived from the bovine N-ERMAD binding protein are indicated by thin lines. The protein cofactor and TKA-1 sequence data are available from EMBL/GenBank/DDBJ under accession numbers U19815 and Z50150, respectively.
Figure 1
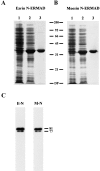
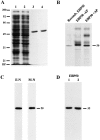
Purification and characterization of ezrin and moesin N-ERMADs. (A and B) show the purifications. Samples were run on a 15% SDS gel and stained with Coomassie blue. Lane 1, total extract of uninduced bacteria; lane 2, total extract of induced bacteria; lane 3, purified proteins. (C) The recombinant proteins exhibit ERMAD activity. Blot overlays using biotinyl ezrin N-ERMAD (E-N) and biotinyl moesin N-ERMAD (M-N) probes on a mixture of ezrin and moesin are shown. The mobilities of molecular mass standards and of placental ezrin (81) and moesin (77) are indicated in kD. DF, dye front.
Figure 2
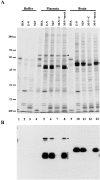
Identification of N-ERMAD binding proteins. (A) Lysis buffer, or placental extracts, or brain extracts were mixed with BSA–agarose (BSA), ezrin–N-ERMAD-agarose (E-N), or moesin– N-ERMAD-agarose (M-N), and then were washed extensively in 0.5 M NaCl and bound proteins eluted and resolved on a 6–20% silver stained gradient SDS gel. For some reactions, a fourfold molar excess of competitor ezrin N-ERMAD (+ C) or mock competitor BSA (+ mock) was added to the extract before mixing with moesin–N-ERMAD-agarose. (B) One quarter the amount of the same samples shown in A were resolved on a 10% gel, transferred to PVDF, and probed with biotinylated ezrin– N-ERMAD. Brackets indicate the N-ERMAD binding proteins, and arrowheads indicate ezrin. The mobilities of standard proteins are indicated in kD. DF, dye front.
Figure 3
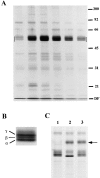
Isolation and characterization of the human placental N-ERMAD binding candidates. (A) An affinity binding assay similar to that shown in Fig. 2, was scaled up 100-fold, and bound proteins were eluted with urea. A silver-stained 12% gel of the peak fractions is shown; the region in which the binding proteins migrate is bracketed. (B) Enlarged view to show resolution of the placental candidates into three species: α, β, and γ. (C) Binding protein heterogeneity is due to phosphorylation. The proteins were recovered from ezrin–N-ERMAD agarose beads by elution with NaI, treated with alkaline phosphatase, and then analyzed on a 10% gel. Lane 1, untreated sample; lane 2, phosphatase-treated sample; lane 3, phosphatase-treated sample in the presence of phosphatase inhibitors. The arrow indicates the migration position of alkaline phosphatase. DF, dye front.
Figure 6
Alignment of the two PDZ domains of human EBP50, rabbit cofactor and human TKA-1 with PDZ domains of selected other proteins: Drosophila Dlg-A (these sequence data are available from EMBL/GenBank/DDBJ under accession number M73529), human chapsyn-110 (accession number U32376), murine PSD-95 (accession number D50621), and human PTPL1 (accession number X80289). The domains have been aligned to optimize the conserved residues, shown in bold, as proposed by Ponting and Phillips (1995). A more complete alignment of other PDZ domains by Ponting and Phillips can be found at:
http://biop.ox.ac.uk/www/dhr.html
Figure 7
Expression, purification, and characterization of recombinant EBP50. (A) Expression and purification of EBP50. Samples were run on an 11.5% SDS gel and stained with Coomassie blue. Lane 1, total extract of uninduced bacteria; lane 2, total extract of induced bacteria; lane 3, EBP50 purified on immobilized ezrin N-ERMAD; lane 4, EBP50 purified on immobilized moesin N-ERMAD. (B) Recombinant EBP50 comigrates with phosphatase-treated EBP50 (+AP) and migrates faster than untreated EBP50 (−AP). The latter two lanes are the same as those shown in Fig. 3_C._ (C) Biotinylated ezrin N-ERMAD (E-N) or moesin N-ERMAD (M-N) binds to recombinant EBP50 by blot overlay. (D) Biotinylated recombinant EBP50 binds to ezrin and moesin N-ERMADs by blot overlay. Samples in panels C and D were resolved on 10 and 15% SDS gels, respectively, and blotted as in Fig. 2. The migration positions of the isolated N-ERMADs (35 kD) and of EBP50 (50 kD) are shown. DF, dye front.
Figure 8
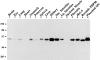
Immunoblot of EBP50 in tissues and cells. 25 μg of total proteins from murine tissues, human placenta and JEG-3 cells, 10 ng of recombinant EBP50, and 2 μg of total proteins of isolated human placental microvilli were resolved on a 11.5% gel, transferred to PVDF, and probed with affinity-purified antibody to EBP50.
Figure 9
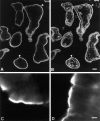
Localization of EBP50 in tissues. In human placenta (A and B), EBP50 (A) colocalizes with actin (stained with rhodamine phalloidin in B) in the microvilli-rich apical regions (arrowheads) of the syncytiotrophoblast. In groups of murine intestinal epithelial cells (C and D), EBP50 (C) is highly concentrated in the microvilli of brush borders, which are also rich in ezrin (D). Bars: (A and B) 20 μm; (C and D) 5 μm.
Figure 10
Localization of EBP50 (A) and ezrin (B) in human JEG-3 cells. The plane of focus was adjusted to the microvilli-rich apical surface of the cells. Bar, 10 μm.
Figure 11
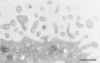
Immunoelectron microscopic localization of EPB50 in human placental syncytiotrophoblast. Note the association of EPB50 specifically with the membrane of the abundant microvilli. Bar, 0.5 μm.
Figure 12
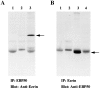
Coimmunoprecipitation of EBP50 and ezrin from human placental microvilli. Extracts of isolated microvilli were subjected to immunoprecipitation with antibodies to EBP50 (A) and ezrin (B). Lanes 1 show a control with antibody but no extract; lanes 2 show a control with extract but no specific antibody; and lanes 3 show the complete reaction. The immunoprecipitates were resolved on a 10% gel, transferred to PVDF, and then probed with biotinylated ezrin antibody (A) or biotinylated EBP50 antibody (B), followed by avidin-peroxidase. In B, lane 4, excess ezrin was added to the lysate immediately before immunoprecipitation. The migration positions of ezrin (A) and EBP50 (B) are indicated by arrows.
Similar articles
- Identification of EPI64, a TBC/rabGAP domain-containing microvillar protein that binds to the first PDZ domain of EBP50 and E3KARP.
Reczek D, Bretscher A. Reczek D, et al. J Cell Biol. 2001 Apr 2;153(1):191-206. doi: 10.1083/jcb.153.1.191. J Cell Biol. 2001. PMID: 11285285 Free PMC article. - Ezrin-radixin-moesin (ERM)-binding phosphoprotein 50 organizes ERM proteins at the apical membrane of polarized epithelia.
Morales FC, Takahashi Y, Kreimann EL, Georgescu MM. Morales FC, et al. Proc Natl Acad Sci U S A. 2004 Dec 21;101(51):17705-10. doi: 10.1073/pnas.0407974101. Epub 2004 Dec 10. Proc Natl Acad Sci U S A. 2004. PMID: 15591354 Free PMC article. - Regulation of cortical structure by the ezrin-radixin-moesin protein family.
Bretscher A. Bretscher A. Curr Opin Cell Biol. 1999 Feb;11(1):109-16. doi: 10.1016/s0955-0674(99)80013-1. Curr Opin Cell Biol. 1999. PMID: 10047517 Review. - ERM-Merlin and EBP50 protein families in plasma membrane organization and function.
Bretscher A, Chambers D, Nguyen R, Reczek D. Bretscher A, et al. Annu Rev Cell Dev Biol. 2000;16:113-43. doi: 10.1146/annurev.cellbio.16.1.113. Annu Rev Cell Dev Biol. 2000. PMID: 11031232 Review.
Cited by
- Ezrin is required for the functional regulation of the epithelial sodium proton exchanger, NHE3.
Hayashi H, Tamura A, Krishnan D, Tsukita S, Suzuki Y, Kocinsky HS, Aronson PS, Orlowski J, Grinstein S, Alexander RT. Hayashi H, et al. PLoS One. 2013;8(2):e55623. doi: 10.1371/journal.pone.0055623. Epub 2013 Feb 6. PLoS One. 2013. PMID: 23405179 Free PMC article. - The ezrin metastatic phenotype is associated with the initiation of protein translation.
Briggs JW, Ren L, Nguyen R, Chakrabarti K, Cassavaugh J, Rahim S, Bulut G, Zhou M, Veenstra TD, Chen Q, Wei JS, Khan J, Uren A, Khanna C. Briggs JW, et al. Neoplasia. 2012 Apr;14(4):297-310. doi: 10.1593/neo.11518. Neoplasia. 2012. PMID: 22577345 Free PMC article. - MCC is a centrosomal protein that relocalizes to non-centrosomal apical sites during intestinal cell differentiation.
Tomaz LB, Liu BA, Meroshini M, Ong SLM, Tan EK, Tolwinski NS, Williams CS, Gingras AC, Leushacke M, Dunn NR. Tomaz LB, et al. J Cell Sci. 2022 Nov 1;135(21):jcs259272. doi: 10.1242/jcs.259272. Epub 2022 Oct 28. J Cell Sci. 2022. PMID: 36217793 Free PMC article. - Rescue of internalization-defective platelet-activating factor receptor function by EBP50/NHERF1.
Dupré DJ, Rola-Pleszczynski M, Stankova J. Dupré DJ, et al. J Cell Commun Signal. 2012 Dec;6(4):205-16. doi: 10.1007/s12079-012-0175-1. Epub 2012 Aug 10. J Cell Commun Signal. 2012. PMID: 22878922 Free PMC article. - Listeria monocytogenes exploits ERM protein functions to efficiently spread from cell to cell.
Pust S, Morrison H, Wehland J, Sechi AS, Herrlich P. Pust S, et al. EMBO J. 2005 Mar 23;24(6):1287-300. doi: 10.1038/sj.emboj.7600595. Epub 2005 Feb 24. EMBO J. 2005. PMID: 15729356 Free PMC article.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Amieva MR, Wilgenbus KK, Furthmayr H. Radixin is a component of hepatocyte microvilli in situ. . Exp Cell Res. 1994;210:140–144. - PubMed
- Berryman M, Franck Z, Bretscher A. Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J Cell Sci. 1993;105:1025–1043. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials