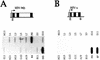Intergenic transcription and transinduction of the human beta-globin locus - PubMed (original) (raw)
Intergenic transcription and transinduction of the human beta-globin locus
H L Ashe et al. Genes Dev. 1997.
Abstract
We have identified novel nuclear transcripts in the human beta-globin locus using nuclear run-on analysis in erythroid cell lines and in situ hybridization analysis of erythroid tissue. These transcripts extend across the LCR and intergenic regions but are undetectable in nonerythroid cells. Surprisingly, transient transfection of a beta-globin gene (epsilon, gamma, or beta) induces transcription of the LCR and intergenic regions from the chromosomal beta-globin locus in nonerythroid cell lines. The beta-globin genes themselves, however, remain transcriptionally silent. Induction is dependent on transcription of the globin gene in the transfected plasmid but does not require protein expression. Using in situ hybridization analysis, we show that the plasmid colocalizes with the endogenous beta-globin locus providing insight into the mechanism of transinduction.
Figures
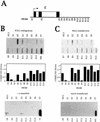
Figure 1
Transcription of the ε-globin gene. (A) Diagram of the ε-globin gene, with the exons (▪), introns (□), and position of the NRO probes marked. (B) NRO analysis of ε-globin transcription in K562 cells in the presence and absence of α-amanitin. The probes used are indicated above and below the filters, and M13 ssDNA serves as a background hybridization control. The probe HIS contains histone DNA and is a positive control for Pol II transcription, whereas the 5S probe contains 5S rDNA and measures Pol III transcription. The signals obtained from the NRO performed in the absence of α-amanitin were corrected for background hybridization and the number of U residues (see Materials and Methods), and plotted on the graph. (C) NRO analysis of ε-globin transcription in HeLa cells following transient transfection of the plasmid εSV−. The corrected signals of the flanking region probes are shown graphically. The control filter shows the NRO result from mock transfected cells.
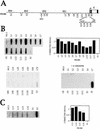
Figure 2
Transcription of the LCR in K562 cells. (A) Diagram of the NRO probes used to detect transcription upstream of the ε-globin gene. The probes shown detect transcription in the direction of the ε-globin gene whereas the equivalent sense probes, denoted by the suffix S, detect antisense transcription in the opposite direction. (B) NRO analysis of LCR transcription in K562 cells in the absence and presence of α-amanitin. The corrected signals from the NRO performed in the absence of α-amanitin are shown graphically, with the exception of probe L4 that contains an Alu repeat. Analysis of transcription by use of sense probes is also shown. (C) NRO analysis of transcription in the 5′-flanking region of the ε-globin gene. The NRO data obtained from K562 cells by use of the shorter LCR probes, L13–L16, are shown with a graph of the corrected signals. The filter has been spliced to remove irrelevant probes.
Figure 7
Transient transfection of a β-globin gene induces all intergenic transcripts. (A) NRO analysis of transcription from the β-globin locus in the nonerythroid cell lines HeLa, 293, and CAK8. These cell lines were transiently transfected with a plasmid containing the ε-globin gene fused to the HIV promoter. A diagram of this construct is shown with the HIV promoter drawn as a shaded box. The HIV-ε plasmid was transfected alone (− tat) or with a Tat-producing plasmid (+ tat). The positions of the probes used relative to the ε, γ, and β genes are shown in previous figures. (B) NRO analysis of LCR transcription in HeLa cells transiently with HIV-ε and a Tat-producing plasmid. The positions of the probes are shown on a diagram of the DNA upstream of ε globin and the NRO data for these probes are shown below. (C) NRO analysis of chromosomal β-globin locus transcription in HeLa cells transiently transfected with ASV−.
Figure 7
Transient transfection of a β-globin gene induces all intergenic transcripts. (A) NRO analysis of transcription from the β-globin locus in the nonerythroid cell lines HeLa, 293, and CAK8. These cell lines were transiently transfected with a plasmid containing the ε-globin gene fused to the HIV promoter. A diagram of this construct is shown with the HIV promoter drawn as a shaded box. The HIV-ε plasmid was transfected alone (− tat) or with a Tat-producing plasmid (+ tat). The positions of the probes used relative to the ε, γ, and β genes are shown in previous figures. (B) NRO analysis of LCR transcription in HeLa cells transiently with HIV-ε and a Tat-producing plasmid. The positions of the probes are shown on a diagram of the DNA upstream of ε globin and the NRO data for these probes are shown below. (C) NRO analysis of chromosomal β-globin locus transcription in HeLa cells transiently transfected with ASV−.
Figure 3
Transcription of the γ-globin gene. (A) Diagram of the NRO probes used to detect γ-globin transcription. Probes with the prefix AG (shaded boxes) contain DNA that hybridizes transcripts from both γ genes to the same extent. Probes shown as solid boxes hybridize transcripts from one γ gene only. The probes indicated by hatched boxes contain equivalent fragments of DNA from each γ gene and there is some cross hybridization to these probes. (B) NRO analysis of transcription of the Aγ gene in HEL cells with the corrected signals shown graphically. (C) NRO analysis of Gγ 3′-flanking region transcription in HEL cells with a graph of the corrected signals. (D) NRO analysis of Aγ transcription following transient transfection of ASV− into HeLa cells. A graph of the corrected signals is shown for the flanking region probes. The NRO results from HeLa cells transiently transfected with pUCA (the test plasmid without the SV40 enhancer) and pUCSV− (the test plasmid without the γ gene) are also shown. In these three transfections, a plasmid containing the adenovirus VAI gene was cotransfected and transcription of this plasmid is detected by the VA probe. (E) NRO analysis of Gγ transcription in HeLa cells transiently transfected with the plasmid GSV− with a graph of the corrected signals.
Figure 3
Transcription of the γ-globin gene. (A) Diagram of the NRO probes used to detect γ-globin transcription. Probes with the prefix AG (shaded boxes) contain DNA that hybridizes transcripts from both γ genes to the same extent. Probes shown as solid boxes hybridize transcripts from one γ gene only. The probes indicated by hatched boxes contain equivalent fragments of DNA from each γ gene and there is some cross hybridization to these probes. (B) NRO analysis of transcription of the Aγ gene in HEL cells with the corrected signals shown graphically. (C) NRO analysis of Gγ 3′-flanking region transcription in HEL cells with a graph of the corrected signals. (D) NRO analysis of Aγ transcription following transient transfection of ASV− into HeLa cells. A graph of the corrected signals is shown for the flanking region probes. The NRO results from HeLa cells transiently transfected with pUCA (the test plasmid without the SV40 enhancer) and pUCSV− (the test plasmid without the γ gene) are also shown. In these three transfections, a plasmid containing the adenovirus VAI gene was cotransfected and transcription of this plasmid is detected by the VA probe. (E) NRO analysis of Gγ transcription in HeLa cells transiently transfected with the plasmid GSV− with a graph of the corrected signals.
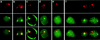
Figure 4
Erythroid tissue primary transcript in situ hybridization. Transgenic mouse fetal liver cells (13.5 day) containing a single copy of the human β-globin locus were hybridized with gene-specific intron oligonucleotides and intergenic transcript probes as follows: (a) human β intron (red), human LCR (green), (b) mouse LCR (red), mouse β-major intron (green), (c) mouse LCR (red), mouse β-major intron and exon (green), (d) human γ 3′ (red), human γ intron (green), (e) human β intron (red), human δ 3′ (green), (f) human β 3′ (red), human β intron (green). Separate red and green images are shown as well as an overlay of the two in the bottom panel.
Figure 5
Transinduction of transcription from the HeLa cell chromosome. (A) NRO analysis of HeLa cells transiently transfected with εSV-ΔFLANK, a plasmid that has the ε 3′-flanking region deleted, as indicated by the crossed-out box. The corrected signals are shown graphically. (B–D) As in A, but the NRO data are from transient transfection of the plasmids GSV–ΔFLANK, ASV–ΔFLANK, and βSV–ΔFLANK, respectively. (D) The position of an Alu repeat in the flanking region of the β gene is marked, and the DNA containing the Alu repeat is not used as a probe. (E) Southern blot analysis of HeLa and HEL DNA. The DNA probe used is equivalent to the NRO probes G10–G14 from the Gγ-flanking region and the position of the probe relative to the two γ genes is indicated on the diagram. This probe has weak homology to the sequences upstream of Gγ because of the γ gene duplication. The fragments predicted by digestion with _Apa_I, _Bam_HI, and _Bgl_II are shown below the diagram. The Southern blot of HeLa and HEL DNA digested with these enzymes is shown with the sizes of the fragments.
Figure 5
Transinduction of transcription from the HeLa cell chromosome. (A) NRO analysis of HeLa cells transiently transfected with εSV-ΔFLANK, a plasmid that has the ε 3′-flanking region deleted, as indicated by the crossed-out box. The corrected signals are shown graphically. (B–D) As in A, but the NRO data are from transient transfection of the plasmids GSV–ΔFLANK, ASV–ΔFLANK, and βSV–ΔFLANK, respectively. (D) The position of an Alu repeat in the flanking region of the β gene is marked, and the DNA containing the Alu repeat is not used as a probe. (E) Southern blot analysis of HeLa and HEL DNA. The DNA probe used is equivalent to the NRO probes G10–G14 from the Gγ-flanking region and the position of the probe relative to the two γ genes is indicated on the diagram. This probe has weak homology to the sequences upstream of Gγ because of the γ gene duplication. The fragments predicted by digestion with _Apa_I, _Bam_HI, and _Bgl_II are shown below the diagram. The Southern blot of HeLa and HEL DNA digested with these enzymes is shown with the sizes of the fragments.
Figure 5
Transinduction of transcription from the HeLa cell chromosome. (A) NRO analysis of HeLa cells transiently transfected with εSV-ΔFLANK, a plasmid that has the ε 3′-flanking region deleted, as indicated by the crossed-out box. The corrected signals are shown graphically. (B–D) As in A, but the NRO data are from transient transfection of the plasmids GSV–ΔFLANK, ASV–ΔFLANK, and βSV–ΔFLANK, respectively. (D) The position of an Alu repeat in the flanking region of the β gene is marked, and the DNA containing the Alu repeat is not used as a probe. (E) Southern blot analysis of HeLa and HEL DNA. The DNA probe used is equivalent to the NRO probes G10–G14 from the Gγ-flanking region and the position of the probe relative to the two γ genes is indicated on the diagram. This probe has weak homology to the sequences upstream of Gγ because of the γ gene duplication. The fragments predicted by digestion with _Apa_I, _Bam_HI, and _Bgl_II are shown below the diagram. The Southern blot of HeLa and HEL DNA digested with these enzymes is shown with the sizes of the fragments.
Figure 6
Further investigation of transinduction. (A) This figure shows the NRO data from HeLa cells transiently transfected with the plasmid GSV–ΔFLANK FS that contains a frameshift mutation in the third exon of the Gγ gene. (B) RNase protection analysis of Gγ transcription in HeLa cells. The riboprobe incorporates the frameshift mutation tested in A and the position of the riboprobe relative to the Gγ gene is indicated. The sizes of the probe fragments protected by the wild-type and mutant Gγ mRNAs are indicated below the diagram. The data show an RNase protection analysis of cytoplasmic RNA isolated from HeLa cells transiently transfected with GSV–FS and GSV−. Specific fragments detected with the riboprobe are labeled A (frameshift) and B (wild-type).
Figure 8
Analysis of the specificity of transinduction. (A) NRO analysis of β-globin transcription in HeLa cells transiently transfected with the plasmid HIV–Mβ and a Tat plasmid. HIV–Mβ contains the mouse β-major gene fused to the HIV promoter as shown. The probes tested have been described. (B) The NRO data obtained from HeLa cells transiently transfected with the plasmid HIV-α and a Tat-producing plasmid are shown.
Figure 9
Plasmid distribution visualized by in situ hybridization. Plasmid and chromosomal transcripts in HeLa cells transiently transfected with the plasmid ASV− were detected with γ gene-specific intron oligonucleotides (green) and either (a) an LCR probe (red) or (b) an ε 3′ probe (red). (c) Detection of plasmid DNA (red) and the human β-globin locus (green) in DAPI-stained nuclei (blue) of HeLa cells transiently transfected with the plasmid ASV−. (d) Detection of plasmid DNA (red) in conjunction with a probe specific to chromosome 22 (green). (a–d) Separate red and green images are shown with the overlay in the middle.
References
- Allan M, Lanyon WG, Paul J. Multiple origins of transcription in the 4.5 kb upstream of the ε-globin gene. Cell. 1983;35:187–197. - PubMed
- Aronow BD, Lattier D, Silbiger R, Dusing M, Hutton J, Jones G, Stock J, McNeish J, Potter S, Witte D, Wiginiton D. Evidence for a complex regulatory array in the first intron of the human adenosine deaminase gene. Genes & Dev. 1989;3:1384–1400. - PubMed
- Ashe MP, Griffin P, James W, Proudfoot NJ. Poly(A) site selection in the HIV-1 provirus: Inhibition of promoter proximal polyadenylation by the downstream major splice donor site. Genes & Dev. 1995;9:3008–3025. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources