OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans - PubMed (original) (raw)
Comparative Study
OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans
H A Colbert et al. J Neurosci. 1997.
Abstract
Although cyclic nucleotide-gated channels mediate sensory transduction in olfaction and vision, other forms of sensory transduction are independent of these channels. Caenorhabditis elegans cyclic nucleotide-gated channel mutants respond normally to some olfactory stimuli and to osmotic stimuli, suggesting that these chemosensory responses use an alternative sensory transduction pathway. One gene that may act in this pathway is osm-9, which is required for each of these responses as well as a mechanosensory response to nose touch. osm-9 encodes a protein with ankyrin repeats and multiple predicted transmembrane domains that has limited similarity to the Drosophila phototransduction channels transient receptor potential (TRP) and TRP-like (TRPL). The sequence of OSM-9 and other TRP-like genes reveals a previously unsuspected diversity of mammalian and invertebrate genes in this family. osm-9 is required for the activity of the predicted G-protein-coupled odorant receptor ODR-10, which acts in the AWA olfactory neurons; its similarity to other G-protein-regulated transduction channels suggests that OSM-9 is involved in AWA signaling. osm-9:: GFP fusion genes are expressed in a subset of chemosensory, mechanosensory, and osmosensory neurons. osm-9 also affects olfactory adaptation within neurons that require the cyclic nucleotide-gated channel for olfaction; in these neurons, the gene has a regulatory function and not a primary role in sensory transduction.
Figures
Fig. 1.
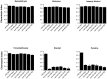
osm-9 mutants do not respond to the AWA-sensed odorants diacetyl and pyrazine. Animals were tested for chemotaxis to a point source of each odorant. Chemotaxis Index (CI) = (number of animals at odorant) − (number of animals at diluent)/(total number of animals). A CI of 1.0 indicates complete attraction; a CI of 0 indicates a random distribution of worms on the assay plate. Each data point represents the average of at least six independent assays using 100–200 animals per assay. Error bars represent SEM. Odorants were diluted in ethanol; 1 μl of diluted odorant was used in each assay. Odorants dilutions were 1:200 benzaldehyde, 1:1000 butanone, 1:100 isoamyl alcohol, 1:1000 2,4,5-trimethylthiazole, 1:1000 diacetyl, and 10 mg/ml pyrazine. AWC responses of osm-9(ky10) were normal across a full range of odorant concentrations (Colbert and Bargmann, 1995; and data not shown).
Fig. 2.
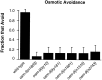
osm-9 mutants do not avoid high osmotic strength. Animals were placed on an agar plate within a 1-cm-high osmotic strength ring consisting of 10–15 μl of 4
m
fructose. Assays were scored after 10 min for retention or escape from the ring. Fraction that Avoid = (number of animals retained by the fructose ring)/(total number of animals assayed). A fraction of 1.0 represents complete osmotic avoidance; a fraction of 0 indicates that all animals escaped the ring. Each data point represents responses of at least 30 animals. Error bars denote the 95% confidence intervals.
Fig. 3.
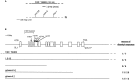
Cloning of osm-9. A, Genetic map position of osm-9 on chromosome IV. The YAC Y44E2 spans the region between unc-33 and_cha-1_ on the physical map. The phage clone λ2–12 contains the osm-9 coding sequence. B, Localization of osm-9 coding sequence. The genomic organization of osm-9 is depicted, along with the location of the n1601 deletion and the_n1603_, n1516, ky10,n2743, and ky161 point mutations.Boxes indicate exons; SL1 refers to the site of attachment of the trans_-splice leader SL1 (Krause and Hirsh, 1987). The YAC Y44E2, the phage λ2–12, and three subclones of the phage were assayed for their ability to rescue the_osm-9 diacetyl chemotaxis defect. Rescue of diacetyl response = (number of independent transformed lines with a diacetyl chemotaxis index >0.4)/(total number of lines assayed). The subclone p[osm-9] also rescued the_osm-9_ nose-touch defects (A. Hart, personal communication; data not shown) and osmotic avoidance defects (Fig.5_E_). Rescuing phage and phage subclones contain ∼1.6 kb of upstream sequence; p[osm-9] contains 3 kb of downstream sequence.
Fig. 4.
Sequence analysis of OSM-9. A, Predicted amino acid sequence of the osm-9 cDNA. Amino acids are numbered beginning at the first methionine. Underlined regions correspond to the three ankyrin motifs found in_osm-9_. Bold underlined regions correspond to the six putative transmembrane domains identified by hydrophobicity analysis. Boxed amino acid residues denote sites of amino acid substitutions in osm-9 mutant alleles. The corresponding substitutions are identified in the_margin_. The amino acid residues deleted in the_n1601_ allele are bracketed by Δs.B, Hydrophobicity plot of OSM-9 derived by Kyte-Doolittle hydropathic analysis (Kyte and Doolittle, 1982). Predicted transmembrane domains are numbered 1 through_6_. C, Comparison of the sixth predicted transmembrane domains of OSM-9,Drosophila TRP (dTRP),Drosophila TRPL (dTRPL), the C. elegans TRP homolog ZC21.2 (cTRP), and three human TRP genes hTRPC1, -2, and -3 (Wes et al., 1995), the predicted C. elegans genes T10B10.7, T01H8.5, and F54D1.5, and human EST zf57d10.s1 (Soares human retina cDNA). Numbers indicate the amino acid residues shown. Residues shared between at least 5 of the 11 genes are_shaded_. The hydrophobic putative transmembrane domain is_underlined_. All of the predicted C. elegans clones have an overall structure reminiscent of TRP, with six hydrophobic domains; the human cDNAs share this structure as far as they extend. D, Sequence alignment of the OSM-9 ankyrin motifs. A comparison of the three OSM-9 ankyrin motifs with the ankyrin consensus motif (designated ANK CON) is shown (Hatada et al., 1992). Numbers indicate the amino acid residue number at the start of each ankyrin motif. Residues identical with those of the consensus are shaded. An_asterisk_ is placed over the consensus glycine residue that is altered in the first and second ankyrin motifs, respectively, in the n1516 and n2743 alleles.
Fig. 5.
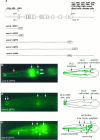
osm-9:: GFP fusion genes are expressed in a subset of sensory neurons. A,osm-9:: GFP fusion genes. Three regions that regulate GFP expression in particular cell types are indicated. All_osm-9:: GFP_ fusion genes were examined in at least three independent transgenic lines. B, A lateral view of the head of a transgenic animal expressing the_osm-9:: GFP2_ fusion gene. GFP is expressed in the four OLQ mechanosensory neurons and in the AWA and ADL amphid chemosensory neurons. All neurons are bilaterally symmetric, and only those on the left side are visible in this focal plane. The fusion protein is present at high levels at the base of the OLQ cilia, but it is only weakly expressed in the cilia themselves.C, A lateral view of the head of a transgenic animal expressing the osm-9:: GFP3 fusion gene, which encodes a GFP-tagged OSM-9 protein. GFP is localized to the OLQ and AWA cilia at the tip of the nose (arrows), but little GFP is present in the cell bodies, axons, or dendrites. ADL expression is weaker or absent in these lines. The fusion gene deletes the last 105 amino acids of OSM-9; when GFP is fused to the last amino acid of OSM-9 (osm-9:: GFP4), a similar but fainter expression pattern is observed. D, A lateral view of the head of a transgenic animal expressing the_osm-9:: GFP5_ fusion gene. GFP expression is present in the OLQ and IL2 sensory neurons and in the AWA, AWC, ASE, ADF, ASG, ASH, ASI, ASJ, ASK, and ADL amphid chemosensory neurons. In more posterior parts of the animal, the FLP, PVD, PHA, and PHB sensory neurons, the ventral uterine cells, and the rectal gland cells also express osm-9:: GFP5. E, Rescue of_osm-9_ chemotaxis and osmotic avoidance by_osm-9:: GFP_ fusion genes. osm-9(ky10); lin-15(n765) animals were transformed with the_lin-15_ plasmid alone (control) or lin-15_plasmid with p[osm-9], osm-9:: GFP3, or_osm-9:: GFP5. Each data point represents 8–24 independent assays. Three independently derived lines of transgenic animals were characterized for osm-9:: GFP3 and six lines for osm-9:: GFP5; all lines gave comparable results (n = 3–4 assays per transgenic line). Error bars represent SEM. Asterisks denote values that are different from the lin-15 control plasmid injection at p < 0.001.
Fig. 5.
osm-9:: GFP fusion genes are expressed in a subset of sensory neurons. A,osm-9:: GFP fusion genes. Three regions that regulate GFP expression in particular cell types are indicated. All_osm-9:: GFP_ fusion genes were examined in at least three independent transgenic lines. B, A lateral view of the head of a transgenic animal expressing the_osm-9:: GFP2_ fusion gene. GFP is expressed in the four OLQ mechanosensory neurons and in the AWA and ADL amphid chemosensory neurons. All neurons are bilaterally symmetric, and only those on the left side are visible in this focal plane. The fusion protein is present at high levels at the base of the OLQ cilia, but it is only weakly expressed in the cilia themselves.C, A lateral view of the head of a transgenic animal expressing the osm-9:: GFP3 fusion gene, which encodes a GFP-tagged OSM-9 protein. GFP is localized to the OLQ and AWA cilia at the tip of the nose (arrows), but little GFP is present in the cell bodies, axons, or dendrites. ADL expression is weaker or absent in these lines. The fusion gene deletes the last 105 amino acids of OSM-9; when GFP is fused to the last amino acid of OSM-9 (osm-9:: GFP4), a similar but fainter expression pattern is observed. D, A lateral view of the head of a transgenic animal expressing the_osm-9:: GFP5_ fusion gene. GFP expression is present in the OLQ and IL2 sensory neurons and in the AWA, AWC, ASE, ADF, ASG, ASH, ASI, ASJ, ASK, and ADL amphid chemosensory neurons. In more posterior parts of the animal, the FLP, PVD, PHA, and PHB sensory neurons, the ventral uterine cells, and the rectal gland cells also express osm-9:: GFP5. E, Rescue of_osm-9_ chemotaxis and osmotic avoidance by_osm-9:: GFP_ fusion genes. osm-9(ky10); lin-15(n765) animals were transformed with the_lin-15_ plasmid alone (control) or lin-15_plasmid with p[osm-9], osm-9:: GFP3, or_osm-9:: GFP5. Each data point represents 8–24 independent assays. Three independently derived lines of transgenic animals were characterized for osm-9:: GFP3 and six lines for osm-9:: GFP5; all lines gave comparable results (n = 3–4 assays per transgenic line). Error bars represent SEM. Asterisks denote values that are different from the lin-15 control plasmid injection at p < 0.001.
Fig. 6.
A C. elegans trp gene is expressed in motor neurons and muscles. A, Expression of ZC21.2:: GFP. Strong expression is present in all RMD, SMD, SMB, RIA, RIB, and RIM motor neurons in the head, many classes of motor neurons in the ventral nerve cord, the BAG sensory neurons, the AVA and SIA interneurons, two pharyngeal neurons, and the vulval and intestinal muscles. No expression was observed in cells that expressed_osm-9:: GFP_ fusion genes.
Similar articles
- Caenorhabditis elegans TRPV ion channel regulates 5HT biosynthesis in chemosensory neurons.
Zhang S, Sokolchik I, Blanco G, Sze JY. Zhang S, et al. Development. 2004 Apr;131(7):1629-38. doi: 10.1242/dev.01047. Epub 2004 Mar 3. Development. 2004. PMID: 14998926 - Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons.
Tobin DM, Madsen DM, Kahn-Kirby A, Peckol EL, Moulder G, Barstead R, Maricq AV, Bargmann CI. Tobin DM, et al. Neuron. 2002 Jul 18;35(2):307-18. doi: 10.1016/s0896-6273(02)00757-2. Neuron. 2002. PMID: 12160748 - Genetics of chemotaxis and thermotaxis in the nematode Caenorhabditis elegans.
Mori I. Mori I. Annu Rev Genet. 1999;33:399-422. doi: 10.1146/annurev.genet.33.1.399. Annu Rev Genet. 1999. PMID: 10690413 Review. - The mechanosensitive nature of TRPV channels.
O'Neil RG, Heller S. O'Neil RG, et al. Pflugers Arch. 2005 Oct;451(1):193-203. doi: 10.1007/s00424-005-1424-4. Epub 2005 May 21. Pflugers Arch. 2005. PMID: 15909178 Review.
Cited by
- What do we know about the transient receptor potential vanilloid 2 (TRPV2) ion channel?
Perálvarez-Marín A, Doñate-Macian P, Gaudet R. Perálvarez-Marín A, et al. FEBS J. 2013 Nov;280(21):5471-87. doi: 10.1111/febs.12302. Epub 2013 May 28. FEBS J. 2013. PMID: 23615321 Free PMC article. Review. - Two forms of learning following training to a single odorant in Caenorhabditis elegans AWC neurons.
Pereira S, van der Kooy D. Pereira S, et al. J Neurosci. 2012 Jun 27;32(26):9035-44. doi: 10.1523/JNEUROSCI.4221-11.2012. J Neurosci. 2012. PMID: 22745502 Free PMC article. - The molecular phylogeny of a nematode-specific clade of heterotrimeric G-protein alpha-subunit genes.
O'Halloran DM, Fitzpatrick DA, McCormack GP, McInerney JO, Burnell AM. O'Halloran DM, et al. J Mol Evol. 2006 Jul;63(1):87-94. doi: 10.1007/s00239-005-0215-z. Epub 2006 Jun 16. J Mol Evol. 2006. PMID: 16786439 - A functional nuclear localization sequence in the C. elegans TRPV channel OCR-2.
Ezak MJ, Ferkey DM. Ezak MJ, et al. PLoS One. 2011;6(9):e25047. doi: 10.1371/journal.pone.0025047. Epub 2011 Sep 21. PLoS One. 2011. PMID: 21957475 Free PMC article. - Genetic and functional diversification of chemosensory pathway receptors in mosquito-borne filarial nematodes.
Wheeler NJ, Heimark ZW, Airs PM, Mann A, Bartholomay LC, Zamanian M. Wheeler NJ, et al. PLoS Biol. 2020 Jun 8;18(6):e3000723. doi: 10.1371/journal.pbio.3000723. eCollection 2020 Jun. PLoS Biol. 2020. PMID: 32511224 Free PMC article.
References
- Aatsinki JT, Lakkakorpi JT, Pietila EM, Rajaniemi HJ. A coupled one-step reverse transcription PCR procedure for generation of full-length open reading frames. Biotechniques. 1994;16:282–288. - PubMed
- Ache BW, Zhainazarov A. Dual second-messenger pathways in olfactory transduction. Curr Opin Neurobiol. 1995;5:461–466. - PubMed
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. - PubMed
- Bargmann CI, Thomas JH, Horvitz HR. Chemosensory cell function in the behavior and development of Caenorhabditis elegans. Cold Spring Harbor Symp Quant Biol. 1990;55:529–538. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases