Mutation of an axonemal dynein affects left-right asymmetry in inversus viscerum mice - PubMed (original) (raw)
Mutation of an axonemal dynein affects left-right asymmetry in inversus viscerum mice
D M Supp et al. Nature. 1997.
Abstract
The development of characteristic visceral asymmetries along the left-right (LR) axis in an initially bilaterally symmetrical embryo is an essential feature of vertebrate patterning. The allelic mouse mutations inversus viscerum (iv) and legless (lgl) produce LR inversion, or situs inversus, in half of live-born homozygotes. This suggests that the iv gene product drives correct LR determination, and in its absence this process is randomized. These mutations provide tools for studying the development of LR-handed asymmetry and provide mouse models of human lateralization defects. At the molecular level, the normally LR asymmetric expression patterns of nodal and lefty are randomized in iv/iv embryos, suggesting that iv functions early in the genetic hierarchy of LR specification. Here we report the positional cloning of an axonemal dynein heavy-chain gene, left/right-dynein (lrd), that is mutated in both lgl and iv. lrd is expressed in the node of the embryo at embryonic day 7.5, consistent with its having a role in LR development. Our findings indicate that dynein, a microtubule-based motor, is involved in the determination of LR-handed asymmetry and provide insight into the early molecular mechanisms of this process.
Figures
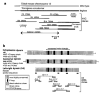
Figure 1
Identification of a dynein gene deleted in lgl. a, YAC contig spanning the mouse DNA/transgene junction in lgl. The location of the newly identified lrd gene (see text) is indicated. b, Homology of cloned lrd sequences with rat cytoplasmic and sea-urchin axonemal dynein heavy chains.
Figure 2
iv has a missense mutation in a highly conserved amino acid of Lrd. a, Raw sequence data. b, The E → K substitution is unique to iv and is in a very highly conserved amino-acid residue (orange). Amino acids that are identical or show only conservative changes across species are shaded. The consensus (bottom) shows residues that are identical among all known axonemal and cytoplasmic dynein sequences, including those not shown here. Amino-acid sequences in this region of Lrd were identical for all wild-type mouse strains analysed (12 in total). The rat orthologue also contains a glutamic acid at this position (data not shown).
Figure 3
RT-PCR analysis of embryonic and adult expression. Primers used were specific for lrd (top) or control β -actin (bottom). Negative controls include: no reverse transcriptase (−RT), no tissue (medium control, −RNA), and no RNA (H2O).
Figure 4
Expression of lrd localized by in situ hybridization. a–d, Hybridization to antisense lrd riboprobe; no signal was detected with a sense control probe. a, c, Bright-field image; b, d, dark-field images. a, b, Sagittal section of an E7.5 embryo demonstrating lrd expression in the node (arrow in b). c, Enlargement to show nodal localization of signal (arrows). d, lrd is expressed in the ciliated epithelium of the adult oviduct (left), but not in the ovary. NR, nodal region; AC, amniotic cavity; PS, primitive streak; AM, amnion; ME, mesoderm; AL, allantois; EE, extraembryonic ectoderm; NE, neural ectoderm. Computer-enhanced colour (yellow) was used to highlight the hybridization signal in c.
References
- Hummel KP, Chapman DB. Visceral inversion and associated anomalies in the mouse. J Hered. 1959;50:9–13.
- Layton WM. Random determination of a developmental process. J Hered. 1976;67:336–338. - PubMed
- McNeish JD, et al. Phenotypic characterization of the transgenic mouse insertional mutation Legless. J Exp Zool. 1990;253:151–162. - PubMed
- Singh G, et al. legless insertional mutation: morphological, molecular, and genetic characterization. Genes Dev. 1991;5:2245–2255. - PubMed
- Lowe LA, et al. Conserved left-right asymmetry of nodal expression and alterations in murine situs inversus. Nature. 1996;381:158–161. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases